INTRODUCTION
During embryonic development, epigenetic mechanisms such as DNA methylation, histone acetylation and non-histone proteins modifications participate in the regulation of gene expression (1, 2). These epigenetic alterations are necessary for the realization of cell differentiation and assembling of two different embryonic compartments, the inner cell mass (ICM) and the trophoectoderm (3).
Two rounds of epigenetic modifications were observed during the mammalian development. Before gametogenesis, the DNA methylation in primordial germ cells is completely erased and subsequently the de novo methylation in male and female occurs. This gender specific remethylation is important for imprinting and erasure of acquired epigenetic modifications (4). After the fertilization in mammalian zygotes (mouse, bovine, porcine, human), the next round of reprogramming begins. The paternal pronucleus is rapidly demethylated, while the maternal pronucleus undergoes passive demethylation (5). The demethylation process continues until the morula stage, when the cells located on the periphery of the embryo are predestined to become a trophoectoderm and the centrally located cells will form the ICM. De novo methylation occurs mainly in the ICM of the blastocyst what correspond with cell differentiation in this layer while the peripheral trophoectoderm cells stay hypomethylated (6).
DNA methylation is catalyzed by specific enzymes, the DNA methyltransferases (DNMTs). Five mammalian DNA methyltransferases, divided into two families, have been identified to date (7). DNA-methyltransferase 1 (DNMT1) is a maintenance enzyme that is responsible for restoring methylation of hemi-methylated CpG dinucleotides after DNA replication (8). Also an oocyte-specific form of this enzyme, DNMT1o, was observed at high concentrations in mature oocytes and early embryos (zygots). Gene targeting experiments indicate that DNMT1o has a role in maintaining methylation marks at maternally imprinted genes in mice (9).
Additional types of the vertebrate cytosine methyltransferase are DNMT3a and DNMT3b. These enzymes catalyze de novo methylation and are thus essential for establishing DNA methylation during development (10).
The aim of this study was to detect the expression of DNMT1 and DNTM3a genes in different stages of embryonic development of bovine vs. porcine intergeneric nuclear transfer (iSCNT) embryos and compare these results with expression in parthenogenetically activated porcine and bovine oocytes. The influence of different ooplasm environment on DNMT1 and DNMT3a genes expression was expected.
MATERIAL AND METHODS
Oocyte collection and in vitro maturation
Bovine oocytes with well developed cumulus cells were isolated from slaughterhouse ovaries of postpubertal Charolais heifers by a slicing method. Selected cumulus-oocyte complexes (COCs) were matured in vitro in tissue culture medium 199 (TCM 199) (Sigma-Aldrich, Germany) containing α-glutamine and 25mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) (Sigma-Aldrich, Germany) supplemented with 22 mg/mL pyruvate, 2.2 mg/mL NaHCO3, 50 mg/mL gentamicin, 10 IU/mL eCG, and 5 IU/mL of hCG (Suigonan, Intervet, Tönisvorst, Germany). Oocytes were in vitromatured in 100 mL maturation medium droplets under silicone oil (Silicone fluid DC200, Serva Biochemica, Heidelberg) for 19h at 39°C and 5% CO2 in humified atmosphere. After maturation, cumulus cells were completely removed by vortexing with 0.1% hyaluronidase in phosphate-buffered saline (PBS). MII oocytes were chosen for further experiments. After maturation, oocytes were prepared for nuclear transfer by completely removing the cumulus-corona by vortexing COCs in 0.1% hyaluronidase (Sigma-Aldrich, Germany) in phosphate-buffered saline (PBS) for 3 min.
Ovaries were collected from prepubertal German Landrace gilts at a local abattoir and transported to the laboratory in physiological saline (0.9% NaCl) solution. Oocytes with at least three complete layers of cumulus cells were collected and matured in vitro for 40 h (11). Then, matured oocytes were transferred to TL-HEPES supplemented with 0.1% hyaluronidase and incubated for 5 min. Cumulus cells were removed by repeated pipetting. Denuded oocytes were transferred to Tyrode’s lactate HEPES (TL HEPES) (Sigma, St. Louis, MO, USA) covered with mineral oil (Silicone fluid DC200, Serva Biochemica, Heidelberg).
iSCNT and parthenogenetic activation
Primary cell cultures were established from bovine (Charolais breed) ear biopsies and female fibroblasts were used as nuclear donors. The cells were cultured in Dulbecco modified Eagle medium-F12 and trypsinized. The harvested cells were resuspended at a concentration of one million cells/mL in freezing medium (10% dimethyl sulfoxide (DMSO; Sigma-Aldrich, Germany), 10% FBS in DMEM) and stored at -70°C. Before transfer into an ooplasm, a suspension of fibroblasts was prepared by standard trypsinization. The cells were pelleted, resuspended and maintained in TCM-air medium.
For production of nuclear transfer embryos MII oocytes were placed in TCM 199 medium enriched with gentamicin, Na-pyruvat, NaHCO3 and BSA (TCM-air), containing 5 mg/mL Hoechst 33342 and 7.5 mg/mL cytochalasin B for 8 min.
Oocytes were enucleated by removing the first polar body and the MII plate. A single fibroblast was transferred into the perivitelline space of the recipient enucleated oocyte. Oocyte-fibroblast cell couplets were electrically fused in a 0.285 manitol based medium containing 0.1 mM MgSO4 and 0.05% bovine serum albumin (BSA) with Multipolator® machine (Eppendorf AG, Germany). Fused cell hybrids, bovine fibroblast in porcine oocyte (iSCNTb), porcine fibroblast in bovine oocyte (iSCNTp) and intact oocytes (parthenogenetic activation) were chemically activated by 5 µM ionomycin. After activation, the embryos were washed and cultured in 30 µL droplets of synthetic oviduct fluid medium supplemented with amino acids and BSA (SOFaa) (Sigma-Aldrich, Germany) supplemented with 0.4% BSA at 39ºC in 5% O2, 5% CO2 and 90% N2 in modular incubation chambers. The embryos were collected at 2, 4, 8 and 16-cell stage for further processing.
RNA extraction and RT-PCR
Total RNA was isolated from pools (triplicates) of 10 embryos each at 2-cell, 4-cell, 8-cell and 16-cell stages derived from iSCNT and parthenogenetic activation using Dynabeads® mRNA DIRECT™ kit (Life Technologies, USA). Subsequently, reverse transcription was carried out using SuperScript First-Strand Synthesis kit (Invitrogen, Carlsbad, USA) in final volume of 20 µl using 2.5 µM random hexamers. Specific primers for bovine and porcine DNMT1 and DNMT3a genes were designed for detection of de novo synthesis of epigenetic enzymes. 18S rRNA was amplified for each embryo to confirm the presence of RNA and as a housekeeping gene for normalization (Tab. 1). As positive controls, porcine and bovine parthenogenetic embryos were used. PCR was performed using Platinum Taq DNA polymerase kit (Invitrogen, Carlsbad, USA) containing MgCl2, dNTP and Platinum Taq DNA polymerase. PCR conditions were as follows: initial denaturation of Taq DNA polymerase at 97°C for 2 min, followed by 35 cycles of 95°C for 15 s, annealing for 30 s (Table 1), and extension at 72°C for 30 s. Amplified PCR products were separated by electrophoresis and visualized using a gel documentation system with EtBr staining and the band intensity was analyzed using ImageJ software (NIH, USA).
Table 1. Species specific primer sequences designed for detection of DNMT1, DNMT3a and 18S rRNA (internal control) genes expression in bovine and porcine embryos. b – bovine specific, p – porcine specific, bp – specific for both species, Ta – annealing temperature
Statistics
Mean values of relative abundances within one embryonic type (iSCNTp, iSCNTb and parthenogenetic) were analyzed by one way analysis of variance method using SigmaStat 3.5 software (Systat Software Inc., San Jose, USA). Significant results from SigmaStat 3.5 were further analyzed by Tukey test. Results were considered significant at P < 0.05. RT-PCR experiments were repeated three times, and data represents the average of all repeats.
RESULTS
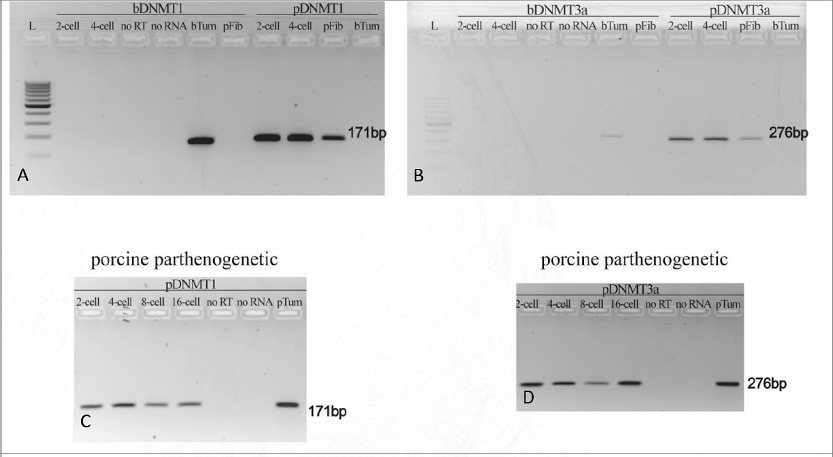
To examine the influence of ooplasm on expression of epigenetic enzyme genes, specific primers for bovine and porcine DNMT1 and DNMT3a genes were designed. The species-dependent specificity of the primers was crucial for the correct determination of the transcript origin (oocytal/somatic). Transcription of bovine DNMT1 (bDNMT1) and DNMT3a (bDNMT3a) genes was not detected in 2-cell and 4-cell stage embryos constructed by bovine fibroblast transfer into the porcine ooplasm (iSCNTb), however, positive results were obtained by applying primers for porcine DNMT1 (pDNMT1) and DNMT3a (pDNMT3a) (Fig. 1A, Fig. 1B). Porcine parthenogenetic embryos were used as positive developmental controls, (Fig. 1C, Fig. 1D).
Figure 1. (A-B) bovine DNMT1 (bDNMT1) (A) and bovine DNMT3a (bDNMT3a) (B) gene expression in comparison with porcine DNMT1 (pDNMT1) (A) and porcine DNMT3a (pDNMT3a) (B) in iSCNT embryos constructed by injecting the bovine fibroblast into the porcine oocyte indicates the lack of de novo transcription of bovine-specific epigenetic enzymes. (C-D) porcine DNMT1 (pDNMT1) (C) and porcine DNMT3a (pDNMT3a) (D) gene expression in porcine embryos after parthenogenetic activation indicates the presence of porcine-specific transcripts of epigenetic enzymes from 2-cell to 16-cell stage
bTum – bovine tumor cells (control of primer specificity)
pFib – porcine fibroblasts (control of primer specificity)
pTum – porcine tumor cells (control of primer specificity)
noRT – no reverse transcriptase in mastermix (negative PCR control)
noRNA – no RNA in mastermix (negative PCR control)
L – 100 bp DNA ladder (Abnova, Taiwan)
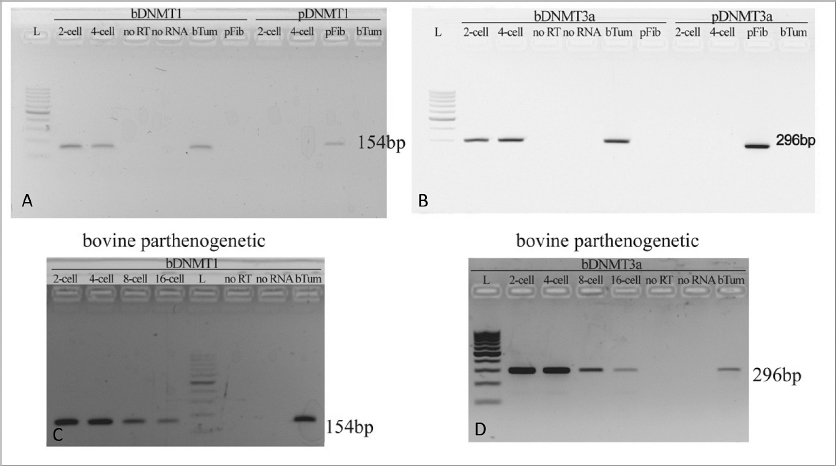
In the 2-cell and 4-cell stage embryos derived from transfer of porcine fibroblast into the bovine ooplasm (iSCNTp) only the primers for bovine DNMTs (bDNMT1, bDNMT3a) showed positive signals (Fig. 2A; Fig. 2B). Considering the different timing of EGA during the embryonic development in bovine and porcine embryos, the intense effect of ooplasm on transferred fibroblast was expected. Despite the mRNA presence of DNMT1 and DNMT3a enzyme of oocyte origin, de novo transcription of somatic DNMT1 and DNMT3a genes was not detected and iSCNT embryos did not develop beyond the 4-cell stage. In parthenogenetic embryos, the continuous supplementation of epigenetic enzymes transcripts is maintained by major genome activation including the transcription of DNMT1 and DNMT3a genes (Fig. 2C; Fig. 2D).
Figure 2. (A-B) bovine DNMT1 (bDNMT1) (A) and bovine DNMT3a (bDNMT3a) (B) gene expression in comparison with pig DNMT1 (pDNMT1) (A) and pig DNMT3a (pDNMT3a) (B) in iSCNT embryos constructed by injecting the porcine fibroblast into the bovine oocyte indicates the lack of de novo transcription of porcine-specific epigenetic enzymes. (C-D) bovine DNMT1 (pDNMT1) (C) and bovine DNMT3a (pDNMT3a) (D) gene expression in bovine embryos after parthenogenetic activation indicates the presence of bovine-specific transcripts of epigenetic enzymes from 2-cell to 16-cell stage
bTum – bovine tumor cells (control of primer specificity)
pFib – porcine fibroblasts (control of primer specificity)
noRT – no reverse transcriptase in mastermix (negative PCR control)
noRNA – no RNA in mastermix (negative PCR control)
L – 100 bp DNA ladder (Abnova, Taiwan)
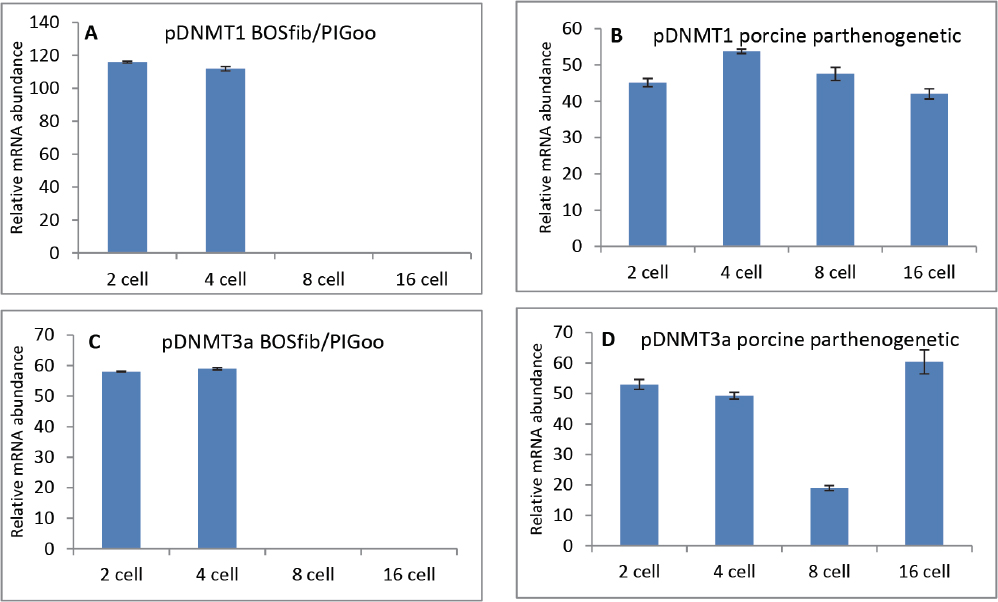
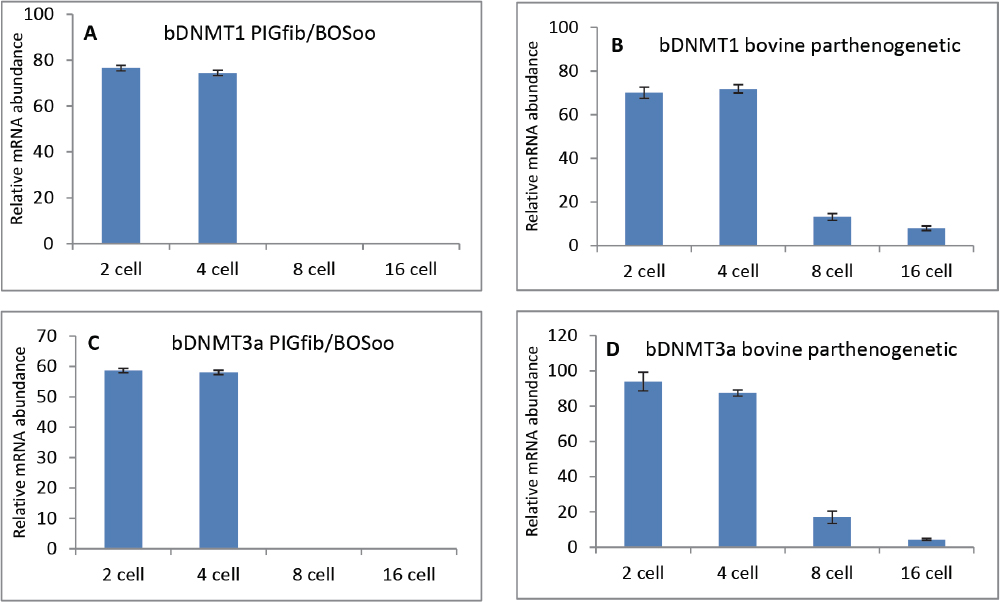
The progress in relative abundance (RA) of the gene transcripts at the 2-cell, 4-cell, 8-cell and 16-cell stages in embryos derived from iSCNT and parthenogenetic activation of bovine and porcine oocytes (PA) is shown in figures 3 A-D and 4 A-D.
Figure 3. Progress in relative abundance of DNMT1 (A, B) and DNMT3a (C, D) transcripts in embryos derived from iSCNT (bovine fibroblast in porcine oocyte) compared with porcine parthenogenetic embryos. mRNA from pools (triplicates) of 10 embryos each at the 2-cell, 4-cell, 8-cell and 16-cell stages were reverse transcribed, and subjected to PCR using species-specific primers. Data are presented as mean ± SEM (bars) of triplicate determinations
Figure 4. Progress in relative abundance of DNMT1 (A, B) and DNMT3a (C, D) transcripts in intergeneric embryos (porcine fibroblast in bovine oocyte) compared with bovine parthenogenetic embryos. mRNA from pools (triplicates) of 10 embryos each at the 2-cell, 4-cell, 8-cell and 16-cell stages were reverse transcribed, and subjected to PCR using species-specific primers. Data are presented as mean ± SEM (bars) of triplicate determinations
The DNMT1transcript level in iSCNTb embryos slightly decreased between 2-cell and 4-cell stages which is in contrast with the progress of RA in porcine PA embryos. The up-regulation of DNMT1 gene in porcine PA embryos was significant (P<0.05) and reached its maximum at the 4-cell stage which corresponds with the major genome activation timing (Fig. 3A-B).
The RA of DNMT3a in iSCNTb embryos was on the same level at the 2-cell and 4-cell stage. The same stages in PA embryos indicated a weak down-regulation of DNMT3a gene towards the 4-cell stage and strong decrease of RA at the 8-cell stage (P<0.001). In accordance with the role of DNMT3a enzyme in de novo methylation process, the increased expression of the corresponding gene was detected at the 16-cell stage of porcine PA embryos (Fig. 3 C-D).
During the 2-cell and 4-cell stage of iSCNTp and bovine PA embryos, no significant differences in expression of any of the DNMT1 and DNMT3a transcripts can be discerned (Fig. 4 A-D). EGA occurs at the 8-cell stage during bovine embryonic development and thus constant RA measurements at the 2-cell and 4-cell stage are likely reflective of maternal transcripts. During the 8-cell and 16-cell stage, a strong down-regulation in the expression of DNMT1 and DNMT3a was observed (P<0.001). Taken together, these results suggest that subsequent to embryonic genome activation, the DNMT1 and DNMT3a genes transcription is silenced, likely as a response to the insufficient demethylation of their promoter domains (12).
DISCUSSION
DNA methyltransferase 1 (DNMT1) is a very intensively studied DNA methyltransferase and is considered to have the most important role in the maintenance of the DNA methylation level following the DNA replication and cell division (13). There are two DNMT1 isoforms present in fully-grown oocytes and preimplantation embryos: oocyte-specific (DNMT1o) and somatic (DNMT1s) isoforms (14). The abundance of the DNMT1o enzyme is very high in nuclei of growing oocytes, however the localization of DNMT1 enzymes in early embryos is strictly cytoplasmatic (15). The ooplasmatic isolation of both enzymes, DNMT1 and DNMT1o, plays a key role in the passive demethylation of the maternal genome in preimplantation embryos. Immunofluorescense studies in mouse and cow have shown the trafficking of DNMT1 from the cytoplasm of 2-cell and 4-cell embryos into the nucleus at the 8-cell stage (4, 9). At the 16-cell stage, DNMT1 enzymes are localized only in the cytoplasm. The timing of the DNMT1 migration into the nucleus coincides with the major genome activation, suggesting that the DNMT3a and DNMT3b genes are activated at the same time.
DNMT3a and DNMT3b are responsible for de novo DNA methylation during development (16). Mechanisms of methylation in different mammalian species are strictly conserved and depend on the expression level of the corresponding genes during oocyte maturation and early embryonic development (17). The influence of somatic cell nuclear transfer (SCNT) on expression of numerous genes in mammals was analyzed and the close linkage between incomplete DNA methylation, as a result of aberrantly expressed DNMT genes, and SCNT success rate was reported (4, 18). Expression of DNMTs genes in bovines was continually detected from the 2-cell stage to the blastocyst stage produced in vitro (19), but the relative abundance of the Dnmt1 mRNA considerably varied between in vivo and in vitro produced bovine embryos. In the in vivo produced embryos DNMT1 gene was significantly less expressed, compared with in vitro derived embryos (20). Likewise, a significant increase in DNMT1 gene transcription at the 8-cell stage was observed in porcine fetal fibroblast nuclear transfer (NT) embryos in comparison with either in vivo produced or fetal porcine skin originated sphere stem cell NT embryos (21). Similar results were obtained in the present study in porcine embryos derived from parthenogenetic activation. However, the DNMT1 gene expression increased earlier, at the 4-cell stage, and this expression level remained until the 8-cell stage. Previous studies have shown that the relative abundance of DNMT1 transcripts in NT porcine embryos was significantly higher compared with IVF embryos and a low level of DNMT1 transcription was observed for in vivo derived embryos (22). DNMT3a mRNA was detected at all stages of porcine embryos with a significant increase in abundance between the 8-cell and morula to blastocyst stages (21) which corresponds with our results and also with the theory of initiation of cell differentiation processes at this stage.
Intergeneric SCNT has been considered as a very effective technique for studying the influence of ooplasm on epigenetic reprogramming of introduced genome which subsequently leads to different aberrations during the embryogenesis. Therefore, the birth of live offspring from iSCNT derived embryos was not reported, and embryonic development after the stage of major embryonic genome activation is strongly hindered (23). Remodeling and reprogramming of the transferred genome is essential for successful embryonic development following SCNT (24).
The presented study is focused on the influence of different ooplasms (porcine and bovine) on the activation of de novo synthesis of DNMT1 and DNMT3a mRNA. It is already known that the matured mammalian oocytes are equipped with efficient amount of epigenetic enzymes and their transcripts required for initial epigenetic processes during early embryogenesis (17). However, these enzymatic supplies are not sufficient for complete reprogramming of transferred nuclei during SCNT, which results in embryonic development breakdown. It is unclear whether the reduced cloning efficiency is related to the inability of a somatic chromatin to undergo the normal changes in methylation pattern as indicated by increased levels of DNMT1 in porcine embryos or to the lack of de novomethylation conditioned by low DNMT3a expression (22). To distinguish the source of mRNA of epigenetic enzymes (ooplasmic and de novo synthesized), intergeneric nuclear transfer (pig vs. bovine and vice versa) was applied in combination with species specific RT-PCR primers detecting DNMT1 and DNMT3a transcription. Our results significantly show the inability of introduced somatic nucleus to initiate and establish the continual expression of observed genes in the environment of ooplasm of different origin. Subsequent to embryonic genome activation, the DNMT1 and DNMT3a genes transcription is silenced, likely as a response to the insuffitient demethylation of their promoter domains (12). This malfunction has led to embryonic development breakdown at the 4-cell stage.
CONCLUSION
The mammalian zygote is able to develop up to the 4-cell or 8-cell embryonic stage (species specific) without major genome activation. The proper cell division is maintained and controlled by maternally inherited factors and under normal conditions is followed by activation of transcription. In the present study the ooplasm with all engaged factors interact with the transcriptionally silenced somatic cell nucleus of different genus. Successful activation of transcription is highly dependent on the correct epigenetic reprogramming of the introduced genome, wherein the epigenetic enzymes, such as DNMT1 and DNMT3s, play a key role. In conclusion, the present study indicates that these enzymes and their epigenetic activities are strictly species-dependent and maternally controlled processes and thus they failed to maintain the proper development of intergeneric SCNT embryos.