INTRODUCTION
Staphylococcus spp. are Gram-positive cocci arranged typically in clusters. The majority of them are facultative anaerobes and catalase-positive. Only two species, Staphylococcus aureus (S. aureus) subsp. anaerobius and Staphylococcus saccharolyticus, are anaerobes and might be catalase-negative (1).
Among the forty-three staphylococcal species that have been described so far, seven species are known to exhibit coagulase-positive or variable reactions: S. aureus, S. intermedius, S. schleiferisubsp. coagulans, S. hyicus, S. lutrae, S. delphini and S. pseudintermedius (2). It is accepted that coagulase production correlates well with the pathogenicity of these bacteria and the coagulase-negative staphylococci (CNS) are deemed as “minor pathogens” (3). Staphylococci are commensals in skin and mucous membranes of the upper respiratory and lower urogenital tract in animals and humans (4). Hence, infections caused by them are endogenous but their prolonged environmental storage, despite non-spore forming, suggests indirect infestation as well (5, 6).
S. aureus is one of the principal bacterial pathogens in humans causing mild to severe life-threatening conditions such as skin, soft tissue, wound infections, and septicaemia, urinary tract infections, endocarditis and osteomyelitis as major sequelae (7, 8, 9). S. aureus is also an important pathogen in veterinary medicine. It has been reported as etiological agent of various infections in cattle, especially mastitis (10), in horses (11), dogs (12), cats (13), rabbits (14), poultry (15) and reptiles (16). Schaumburg et al. (17) and Monecke et al. (18) have also isolated S. aureus from non-human primates in Africa and from diseased hedgehogs in Sweden, respectively.
S. aureus, and other coagulase-positive staphylococci produce a wide range of virulence factors, including toxins, enzymes, biofilm and factors that counteract the immune system defense. These determinants are responsible for the pathogenesis of staphylococcal diseases (19). Enterotoxins (A-E) cause food poisoning in humans. SSSS-syndrome in newborns and dogs is due to the exotoxin exfoliatin. Exudative epidermitis in pigs and some skin diseases in humans and dogs are associated with epidermolytic toxins (6). Gangrenous mastitis is due to the α-haemolysin production. The exotoxin, Panton-Valentine leukocidin kills neutrophils and macrophages in cattle, rabbits and humans. The surface protein A of S. aureus blocks the specific binding of IgG class immunoglobulins. Enzymes produced by staphylococci include staphylokinase, coagulase, hyaluronidase, lipase, collagenase, proteases, nucleases and urease (6). Some virulence factors are important laboratory tools in detection of staphylococci. Thus, catalase is an enzyme leading to survival of bacteria in macrophages (20) but also the main conventional test for primary differentiation of staphylococci from streptococci. The tube coagulase test categorises staphylococci into two groups differing in pathogenicity as mentioned above. Staphylococcal nuclease hydrolyses DNA and RNA of host cell and thereby facilitates tissue spread. It is also a unique genetic marker for S. aureus detection in different clinical samples (21).
The aim of the present study was to detect Staphylococcus aureus among other coagulase positive staphylococci from animal origin by using conventional methods (biochemical and latex agglutination tests) and a molecular method, based on the nuc gene, as the gold standard and to assess the usefulness of these methods.
MATERIAL AND METHODS
Bacterial strains
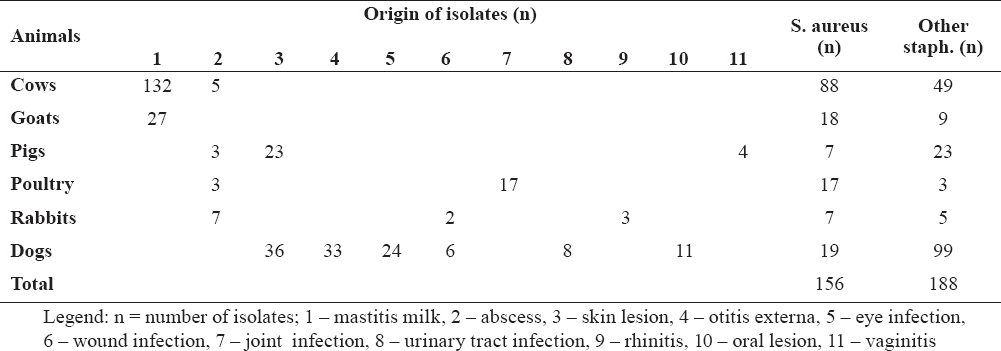
A total of 344 Staphylococcus spp. isolates from cows (n=137), goats (n=27), pigs (n=30), birds (n=20), rabbits (n=12) and dogs (n=118) were included in the study. Isolates originated from various pathological conditions – mastitis milk, abscesses, skin lesions, otitis externa, infections on eyes, wounds, joints, urinary tract, rhinitis, oral cavity lesions and vaginitis. Samples were cultured on tripticase soy blood agar (TSA, Fluka, India supplemented with 5% defibrinated sheep blood) and on MacConkey agar (NCIPD, Sofia, Bulgaria). Plates were incubated at 37°C for 24-48 h under aerobic conditions. S. aureus ATCC 25922 was used as control strain in all experiments.
Biochemical tests
The following biochemical tests and characteristics of bacterial strains were taken into consideration – Gram staining, catalase and oxidase tests, colony pigmentation, haemolysis, tube coagulase test (rabbit plasma supplied by NCIPD, Sofia, Bulgaria), VP test, ONPG (β-galactosidase, 4 mg disk, HiMedia, India), test with polymyxin B (300 unit disk, Oxoid, UK), acid from mannitol determined on mannitol salt agar (NCIPD, Bulgaria), utilisations of trehalose (NCIPD, Sofia, Bulgaria) and maltose (MkB Test, Rosina, Slovak republic). All tests were carried out according to the manufacturer’s instructions and in compliance with the general bacteriology procedures.
Latex agglutination
A Staphylococcus Rapid Latex Kit (Atlas Medical, Cambridge, UK) for the detection of S. aureus in cultures was used following precisely the instruction of the company.
PCR based on nuc gene
DNA was extracted without using a commercial kit. Bacterial suspensions were prepared in sterile distilled water, boiled and centrifuged for 5 min at 12,000 g (22).
The concentration and purity of DNA extracts were determined by DNA/RNA spectrophotometer Gene Quant 1300 at A260/A280. The DNA extracts were stored at - 20°C until the beginning of the trials. PCR was run as described by Brakstad et al. (23) with an expected amplicon size of 270 bp.
RESULTS
Based on the biochemical methods, a total of 156 Staphylococcus aureus strains were presumptively detected among the studied 344 isolates (Table 1). The majority of S. aureus originated from cows with mastitis (n=88), 19 strains were isolated from dogs, 18 from goats and 17 from birds (chickens).
Table 1. Origin of isolates
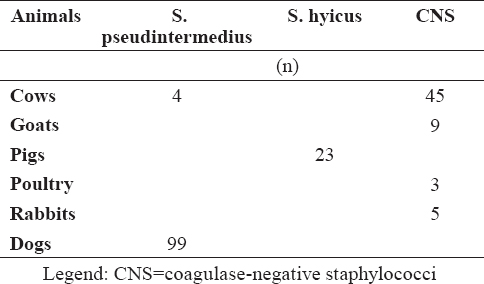
Other staphylococci were determined as S. pseudintermedius (n=103), S. hyicus (n=23) and CNS (n=62) with biochemical tests (Table 2).
Table 2. Distribution of staphylococci other than S. aureus in examined samples
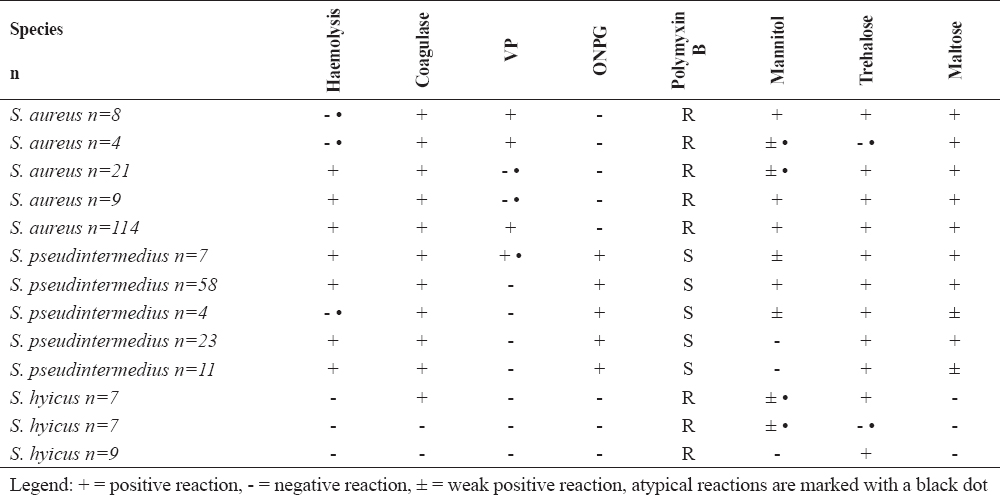
Biochemical profiles of S. aureus and other important coagulase-positive/variable staphylococci are presented in Table 3. Most of the S. aureus strains (114/73%) showed biochemical reactions typical for the species compared to the control strain such as presence of β-haemolysis, positive tube coagulase test, production of acetoin (VP test), negative ONPG test, resistance to polymyxin B, acid from mannitol, trehalose and maltose. A total of 30 strains were VP negative (19.2%), 25 (16%) fermented mannitol weakly and 4 strains did not produce acid from trehalose (2.6%). Twelve strains were found to be non-haemolytic (7.7%). In twenty one strains (13.5%) and in 4 strains (2.6%) two and three atypical reactions were observed, respectively. The majority of S. pseudintermedius(58/56.3%) exhibit species-specific biochemical behaviour - β-haemolysis, positive tube coagulase test, negative VP test, positive ONPG test, susceptibility to polymyxin B, fermentation of mannitol, trehalose and maltose. Four S. pseudintermedius strains were gamma-haemolytic. Twenty three staphylococcal isolates from pigs were differentiated as S. hyicus.
Table 3. Key biochemical tests for clinically relevant staphylococci
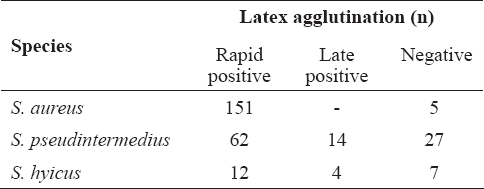
The results from the performed latex agglutination test are shown in Table 4. The test led to rapid positive result (up to 10 s) with almost all S. aureus strains tested with the exception of 5 strains from cows with mastitis (3.2%). Most of S. pseudintermedius strains also gave a rapid agglutination (n=62), 14 strains were positive within 1 min and 27 were negative. Agglutination was observed with some S. hyicus strains as well.
Table 4. Latex agglutination with coagulase positive/variable staphylococci
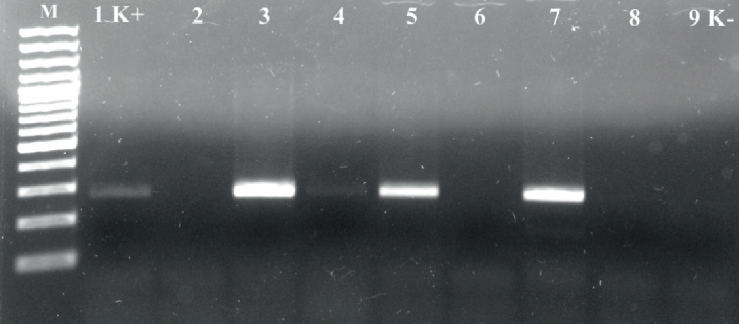
PCR assay performed with strains biochemically identified as S. aureus resulted in amplification of nuc gene fragments with an expected molecular size of 270 bp. Specific amplicons with other coagulase positive/variable staphylococci were not observed (Fig. 1).
Figure 1. PCR for detection of nuc gene in Staphylococcus strains, M – molecular marker, 100 bp; 1 K+ -reference strain; 2, 6 – S. pseudintermedius; 3-5, 7 – S. aureus strains; 8 – S. hyicus; 9 K- - negative control
DISCUSSION
The identification of coagulase-positive staphylococci at the species level is essential for the laboratory diagnostic practice. For instance, the interpretation of minimum inhibiting concentrations of oxacillin for detection of methicillin-resistant staphylococci depends on the Staphylococcus species (2, 24). The erroneous determination of concentrations could influence the antibiotic drug decision making and hence, the therapy of infections. Therefore, the detection of S. aureus among other coagulase-positive staphylococci is an important task for veterinary medicine, as well.
In many laboratories, the S. aureus identification is based on conventional biochemical tests, of which the tube coagulase is the main test for differentiating pathogenic staphylococci (25). According to some authors, the differentiation of coagulase-positive staphylococci based on their phenotypic characteristics is uncertain because of the lack of unique species-specific biochemical markers (26). In this study, we selected key biochemical tests for fast and accurate detection of clinically relevant staphylococci from animal origin (6). The panel included tests with established minimum variations in the behaviour of the common pathogenic staphylococci such as detection of free coagulase, presence of haemolysis, VP test for acetoin production, ONPG test to demonstrate β-galactosidase, test with polymyxin B, utilisation of mannitol, trehalose and maltose. The colony pigmentation was taken into account in primary differentiation of the isolates, as well.
The analysis using biochemical tests has undoubtedly determined 114 strains as S. aureus which exhibited a typical biochemical profile compared to that of the reference S. aureus ATCC 25922 strain and specified in the Clinical Veterinary Microbiology reference book (6). The other 42 strains showed variable behaviour with respect to some parameters and were classified as suspicious for S. aureusuntil the latex agglutination and PCR tests. Acetoin production is one of the main tests for discrimination of S. aureus from S. pseudintermedius and S. hyicus. An interesting finding in the present study was the detection of 30 (19.2%) acetoin-negative S. aureus and 7 (6.8%) VP-positive S. pseudintermedius strains. The strains originated from cows with mastitis and dogs affected by otitis externa, respectively. In a study on the phenotype characteristics of 96 S. aureus isolates from poultry meat in Spain, Capita et al. (27) demonstrated 100% acetoin production. Similar data about acetoin are established by El-Jakee et al. (5) in S. aureus strains from human and animal origin (cattle, dogs, chickens). In the present study, other atypical reactions were observed with regard to fermentation of mannitol, trehalose and to haemolytic activity. Twenty five S. aureus fermented mannitol weakly without alteration of mannitol-salt agar colour into yellow, but producing orange-yellow colonies. These strains were isolated from cows with mastitis. Another finding was the isolation of 4 strains with reactions atypical for S. aureus to three parameters included in the panel: they originated from dogs with skin lesions and were characterized with lack of haemolysis, weak mannitol fermentation and no acid production from trehalose. Furthermore, the colonies of these isolates were non-pigmented but VP positive, ONPG negative, polymyxin B-resistant and maltose-fermenting – tests useful to distinguish S. aureus from other pathogenic staphylococci. Variable responses to ONPG, polymyxin B and maltose were not established which allowed concluding that these tests could be reliable markers in the identification of animal S. aureus isolates. Gandra et al. (28) compared two phenotypic tests – ONPG and sensitivity to acriflavine for differentiation of coagulase-positive S. aureus, S. intermedius and S. hyicus, vs PCR of coa and nuc gene sequences. The authors established similar discriminatory power of both techniques.
Latex agglutination tests for S. aureus detection are based on the presence of clumping factor and/or protein A that differentiate staphylococci which do not possess these factors. Only 5 strains isolated from cow mastitis failed in the test. Similar to our results are the findings of Zschöck et al. (29) who evaluated 6 commercial agglutination kits for S. aureus detection in bovine mastitis. The authors conclude that slide agglutination methods can provide rapid identification of S. aureus from bovine mastitis as well, however the sensitivity and specificity of these tests is lower than those reported for human isolates. Latex agglutination tests may be useful for human S. aureus identification in routine diagnostic purposes (30), but concerning other staphylococcal isolates it has to be taken into account that some coagulase-positive/variable staphylococci such as S. pseudintermedius and S. hyicus, tested in the present study may give rapid agglutination. Therefore, we suggest that these tests alone should not be used for S. aureus detection in veterinary medicine.
All strains identified preliminary as S. aureus were confirmed by PCR based on nuc gene. This gene, coding a thermostable nuclease, seems to be highly specific and a unique genetic marker for S. aureus detection confirmed both in our and previous research (21, 31, 32). However, there is a report about nuc negative S. aureus clade from animals in sub-Saharan Africa (33) harboring a homologue of nuc gene. This clade nuc sequence is highly divergent from those of nuc S. aureus reference strains. Therefore, it gives false-negative results in standard nuc PCR techniques applied alone (33).
Staphylococcus spp. show a certain host specificity as the strains isolated from clinical materials differ from species to species, for instance, prevailing species of isolates from ruminants, pigs and dogs are S. aureus, S. hyicus and S. pseudintermedius, respectively (26, 34), which is in line with our results. Although less frequently, coagulase-positive species other than S. aureus are also involved in the etiology of bovine mastitis – such as S. pseudintermedius and S. schleiferi subsp. coagulans detected in a previous study of ours with BioLog identification system (22). According to Markey et al. (6) S. pseudintermedius, S. schleiferi subsp. coagulans and S. aureus are isolated most commonly from dogs. In the present research, no S. schleiferi subsp. coagulans was found in canine samples, whereas the occurrence of S. aureus from skin lesions and infected wounds was 16.1% (19/118). Comparable results (16%) are reported by El-Jakee et al. (5), about the prevalence of S. aureus in samples from dogs. As the other species included in the present work are concerned, our data was comparable to published results (35).
CONCLUSION
The atypical biochemical behaviour and the absence of clumping factor and/or protein A in some S. aureus strains established in this study suggests that the precise detection of S. aureus from animal origin should be done by combination of conventional and molecular methods. In cases where strains show a typical biochemical profile based on the chosen key tests such as β-haemolysis, tube coagulase test, VP test, ONPG test, resistance to polymyxin B, acid from mannitol, trehalose and maltose, the detection of S. aureus can be done without PCR confirmation. We believe that the results in this study will add to the clinical microbiology experience.
CONFLICT OF INTEREST STATEMENT
The authors declared that they have no potential conflict of interest with respect to the authorship and/or publication of this article.