INTRODUCTION
Salmonella is an enteric, facultative intracellular and ubiquitous pathogen with a high zoonotic potential (1). The species Salmonella enterica is the most important pathogen of the genus and includes six subspecies: enterica, indica, houtenae, salamae, arizonae, diarizonae. The subsp. enterica is usually implicated in warm-blooded host’s infections with major zoonotic importance (2, 3). Infections due to Salmonella may be asymptomatic or may persist through gastrointestinal infections to potentially fatal systemic disease. In the United States as a developed country with very high hygienic surveillance, there are an estimated 1.0 million annual cases of human nontyphoidal salmonellosis with approximately estimated 380 annual deaths and 19,000 estimated annual hospitalizations, while in other regions these figures may be higher due to poor hygienic criteria (4). The death toll is much higher in third-world countries, where typhoid fever is a major killer due to poor sanitary conditions. In Iran, there are thousands of reported cases annually of foodborne diseases related to Salmonella (5, 6). This infectious agent is transmitted from animal reservoirs to humans mainly via contaminated foods, direct/indirect contact with infected hosts or environment.Today, zoonosis with livestock or captured wild life reservoirs constitutes a superior public health problem (7-9).
There are several documents indicating that the prevalence of Salmonella in captured wildlife as well captured species in zoos and petting exhibitions has increased significantly in recent years. Various factors including stress because of food shortage, poor husbandry with overcrowding and the introduction of new hosts may cause the distribution of Salmonella serovars in the environment and the increase in new reservoirs for humans (10-13). Thereare no documented investigations about antimicrobial resistance of Non-Typhoidal salmonella (NTS) strains circulating in wildlife kept in zoo in the Semnan province, but some information is available regarding Salmonella infection and their antimicrobial resistance profile (versus different antibiotics) from wild captive animals in other regions of Iran. For example, in a study carried out within free living sparrows in Tehran suburbs, all the Salmonella isolates were sensitive to Norfloxacin, Flumequine, Ampicillin, and Sultrim, and 35% were resistant to Lincospectin (3). In another study regarding the antimicrobial resistance profile of Salmonella strains from birds in parks and pet shops in Tehran, all isolates were susceptible to Danofloxacin, Norfloxacin, Levofloxacin, Amikacin, Gentamicin, and Tobramycin. Resistance to other antibacterial agents was variable and ranged from 0-57.9% (12). Also, a similar study in the Mashhad zoo (Khorasan-e-Razavi, East of Iran) regarding salmonella antimicrobial resistance phenotypes from wild captive herbivores showed the highest rate of resistance against Amoxicillin (100%), Tetracycline (80%), Neomycin (60%), Lincospectin (50%) and Enrofloxacin (40%), while resistance to Furazolidone wasn’t observed (14). Another report about resistance to Fluoroquinolones within non-typhoidal Salmonella serovars from human cases showed a 6.3% resistance rate (15).
The aim of the present study was to evaluate the prevalence of Salmonella enterica serovars Typhimurium and Enteritidis, the two major serovars implicated in human salmonellosis, within wild birds and mammals kept in captivity in the Semnan zoo and pet shops in Semnan City using biochemical and genotypic assays. Furthermore, we provide the assessment of antimicrobial resistance phenotype of the isolated Salmonella strains.
MATERIAL AND METHODS
Sample collection, isolation of Salmonella strains and serotyping
This is a cross-sectional study carried out in the period from winter to April of 2015-2016 in Semnan City, Semnan Province (north-east of Iran). In total, 152 fecal samples were collected from animals which were healthy (except for two monkeys that were both suffering from diarrhea), clinically. All animals were adult, kept in fences/cages separate from other species without direct contact to visitors in the zoo and the pet shops. The host species and the number of samples from each species (including mammals and birds) were as follows (Taxonomic Name/English Name (No. of animals testes): Macaca mulatta/Rhesus macaque (2); Aquila pomarina/Lesser spotted eagle (10); Tyto alba/Bran owl (2); Equus ferus caballus/Horse (4); Ursus arctos/Brown Bear (2); Capra aegagrushircus/Wild Goat (5); Psittacula krameri/Ring-necked parakeet (15); Numida meleagris/Guineafowl (5); Canis lupus/Wolf (15); Dama dama mesopotamica/Persian fallow deer (1); Oryctolagus Cuniculus/Rabbit (4); Pavo muticus/Green peafowl (4); Carduelis carduelis/Goldfinch (10); Taeniopygia guttata/Zebra finch (10); Serinus canaria/Canary (6); Psittacus erithacus/Congo African grey parrot (4); Acridotheres tristis/Common Myna (8); Phasianus colchicus/Common pheasant (15) and Columba livia/Pigeon (30). Faecal samples were placed on ice and transported immediately to the microbiology laboratory (Faculty of Veterinary Medicine, Semnan University), and Salmonella strains were isolated using standard (Culturing & Biochemical) methods as described by Mirzaei et al. (3). All isolated Salmonella strains were then serotyped using Salmonella H Antisera and Salmonella O Antisera (Difco). Briefly, the Salmonella isolates were cultured onto TSI slant medium and grown overnight at 37 °C, and afterward the isolates were serotyped using antisera O (B, D, E, C) and H, respectively to determine the serogroups and serotypes of the isolates (6, 7).
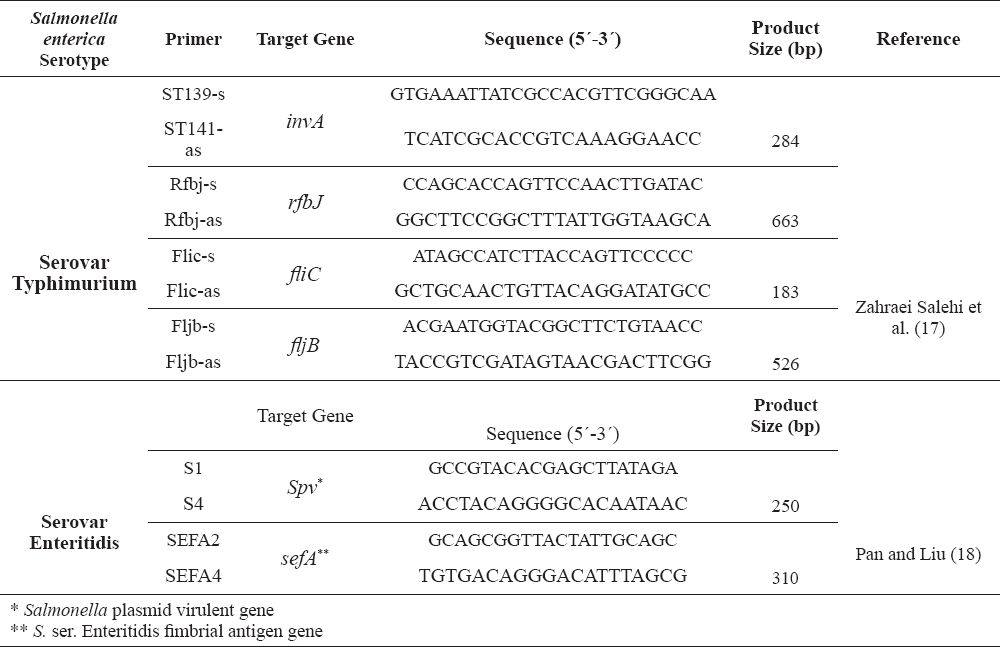
DNA extraction and multiplex-PCR
Genomic DNA was extracted from Salmonella isolates with the Rapid One-Step Extraction (ROSE) method (16). Then, Multiplex PCR was conducted with 2 independent sets for genetic identification of Salmonella serovar Typhimurium (targeting invA, rfbJ, fliC and fljB) and Salmonella serovar Enteritidis (targeting spv and sefA) (primers sequences present in Table 1). For Salmonella ser. Typhimurium, PCR was performed in a 25 μL volume containing 40 ng of total Salmonella ser. Typhimurium extracted DNA, 8 µL of each primer and 13 µL of 2X-PCR master mix (Jena, Takapouzist). Thermal and electrophoresis conditions were as previously described by Zahraei Salehi et al. (17). For Salmonella ser. Enteritidis, PCR was performed in a total volume of 25 µL amplification mixture consisting of 15 µL of 2X mentioned Master mix, 1 µL of each primer and 3 µL of DNA (40 ng) that was extracted as template using thermal and gel electrophoresis condition described by Pan and Liu (18).
Table 1. Primers used for the genetic characterization of Salmonella serovars Typhimurium and Enteritidis
Antimicrobial susceptibility testing
Antimicrobial susceptibility of confirmed Salmonella isolates was determined by the disk diffusion method according to the guidelines provided by the Clinical and Laboratory Standards Institute (CLSI) (19) and twelve antibiotic disks were applied to identify the antimicrobial resistance phenotype of the isolated strains as follow: Ampicillin; Ceftizoxime; Oxytetracycline; Streptomycin, Lincospectin, Neomycin; Imipenem, Meropenem; Nalidixic acid; Furazolidone; Flurefenicole and Chloramphenicole. Finally, distribution of antimicrobial resistance versus two diagnosed serotypes (Enteritidis and Typhimurium) were analyzed using Chi-Square (X2) and Fisher’s Exact Test at 95% of confidence and P = 0.05 of significant level.
RESULTS
Fecal samples were collected from 152 animals (115 birds and 37 mammals), and Salmonella spp. strains were found in 27 (23.4%) and 5 (13.5%) of these animals, respectively.
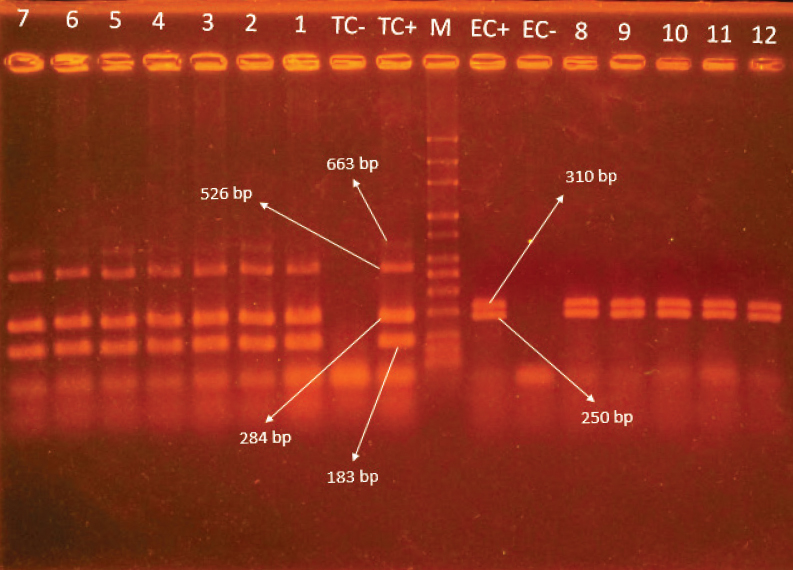
In total, thirty two Salmonella strains were isolated from 152 (21%) fecal droplets of various animals kept in captivity consisting of 19 (12.5%) Salmonella serovar Enteritidis (all strains were isolated from bird’s species) and 13 (8.5%) serovar Typhimurium (5.2% isolates from birds, 2% from mammals and 1.3% from rodents, respectively). Distribution of Salmonella serovars within monitored animals, including the number of isolates from each host, are presented in Table 2. The results of Multiplex-PCR gel electrophoresis for genetic confirmation of Salmonella serovars are present in Figure 1.
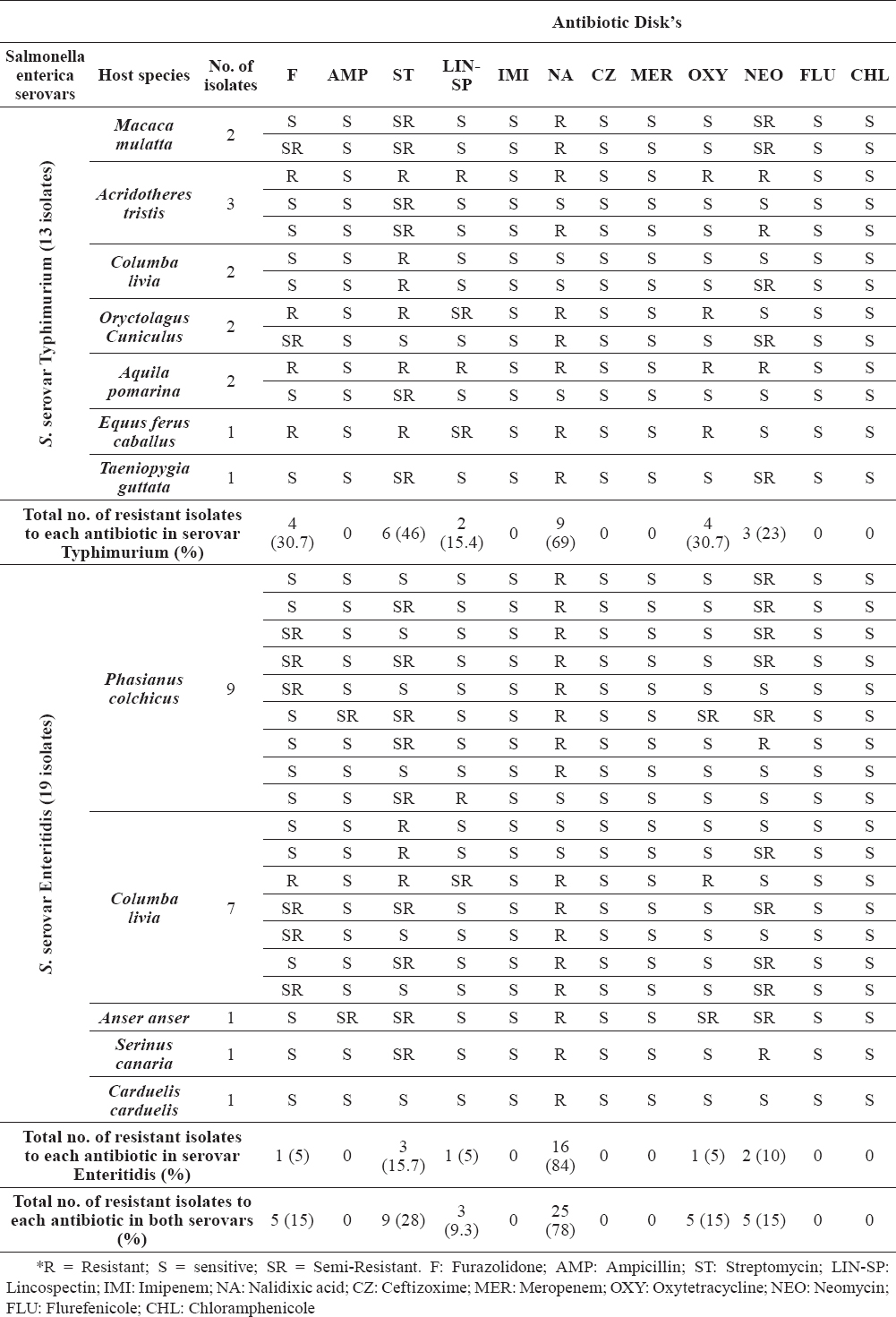
Table 2. Distribution of antibiotic resistance patterns in Salmonella serovars Typhimurium and Enteritidis isolates in this study with the source of isolation using Disk Diffusion method
Figure 1. Gel electrophoresis of Multiplex-PCR for detection of S. serovars Typhimurium & Enteritidis. Lane M: 50 bp marker. Lanes TC- & EC-: negative controls for Typhimurium and Enteritidis, respectively. Lanes TC+ & EC+: positive controls for Typhimurium and Enteritidis serovars, respectively. Lanes 1-7: Typhimurium isolates. Lane 8-12: Enteritidis isolates
The overall majority of the Salmonella isolates (93.7%) showed resistance to at least one of the antibiotics tested except for two Salmonella serovar Typhimurium strains (6.3%), which were sensitive to all antimicrobials. Antimicrobial resistance was observed for six antibiotics including Nalidixic acid (78%) as the highest resistance, then Streptomycin (28%), Oxytetracycline, Neomycin, Furazolidone (each 15%) and Lincospectin (9.3%), respectively and all isolates showed sensitivity to Chloramphenicole, Flurefenicole, Meropenem, Ceftizoxime, Imipenem and Ampicillin antibiotics. The distribution of antimicrobial resistance patterns within all isolates, S. ser. Enteritidis and S. ser. Typhimurium are summarized and presented in Table 2.
Statistical analysis of the results revealed that there is no significant difference in distribution of antimicrobial resistance patterns between the two isolated serotypes. Statistical analysis between isolates of serovars Typhimurium and Enteritidis showed that the resistance to only one antimicrobial in Enteritidis strains are higher than Typhimurium ones (p = 0.014), significantly. There was no significant difference between isolates belonging to both serotypes from a Multidrug-Resistance (Resistance to more than two antibiotics) point of view.
DISCUSSION
Salmonella presence and isolation from captured wild animals has been reported from all over the world and human infection by exotic-associated salmonellosis has been increasing throughout the world because more people are in contact with exotic species not only indoors, but also at zoos and petting exhibitions. (20-24). Although infection of humans with salmonella serovars in zoos is not common, there is some published data demonstrating the outbreaks of non-typhoid Salmonellosis in zoo visitors. For example Friedman et al. (25) reported an outbreak of Salmonellosis in 39 children visiting a reptile exhibition in Denver, Colorado (25) or since 1966 until now, there are several other published zoonotic disease outbreaks associated with animal exhibits (24, 26, 27). Therefore, zoo visitors and zookeepers are at risk of infection from animal carriers. The aim of this study was to ascertain the prevalence of Salmonella serovars Enteritidis and Typhimurium in zoo animals at Semnan Zoo and some pet shops in Iran.
Salmonella serotypes belonging to subspecies I are responsible for more than 99% of Salmonella infections in humans. This is because generally, these members are found in warm blooded hosts and to a lesser extent in cold blooded animals like reptiles, whereas other subspecies are isolated from cold-blooded vertebrates and their environments. All 32 Salmonella isolates in our study belonged to subspecies I (59.3% Salmonella ser. Enteritidis and 40.6% Typhimurium) and according to the Centers for Disease Control and Prevention (CDC), Salmonella serovars Typhimurium and Enteritidis are the two most common serovars associated with human disease, therefore, occurrence of these strains in zoo and pet species in Semnan represents a potential threat to public health (4, 28). In some developed countries like the United States and in other areas of the world, Salmonella serovar Typhimurium is the second most commonly reported serotype causing salmonellosis in humans and the major cause of salmonellosis in wild birds with high mortality among infected birds (29-31). This data suggest the high importance of this serotype in animals mortality kept in captivity in Semnan, especially birds. Furthermore, the serovar Typhimurium has been the most common reported serotype distinguished in other non-human hosts, underscoring the wide host range and complex ecology of this serotype (23). This fact can explain better the prevalence of Typhimurium serovar in exotic species other than birds in the Semnan zoo and pet shops. In the present study, Salmonella serovar Enteritidis was identified as the most prevalent serovar within avian species and this is in parallel with other studies in Iran where the prevalence of Salmonella serovars within domestic/wild and pet birds has been demonstrated (3, 12, 32). Keeping multiple birds in close contact with each other and in contact to other non-bird species provides the ideal environment for infectious agents to spread easily with susceptible hosts. This explains why serovar Typhimurium was another prevalent agent in our study in bird species. In common with this, wildlife can be infected with Salmonella serovars before capturing via contact to livestock or other wild reservoirs. The high prevalence of such serovars within zoo and animal exhibitions can be due to rodents or free living birds like sparrows circulating within species cages/area, as well as the close and intense contact between them (3).
There has been a serious increase in resistance of Salmonella serovars to antibiotics over the last years and territorial variations of resistance to antibiotics may be explained in part by different regional antimicrobial administrations (33, 34). Antimicrobial resistance phenotypes of Salmonella isolates obtained in the present study showed resistance to Nalidixic acid (78%), Streptomycin (28%), Oxytetracycline, Neomycin, Furazolidone (each 15%) and Lincospectin (9.3%), respectively and all isolates showed sensitivity to Chloramphenicole, Flurefenicole, Meropenem, Ceftizoxime, Imipenem and Ampicillin antibiotics. Today, some antimicrobials are not used any more in human practice, such as Furazolidone and Chloramphenicol, but some are still in use in animal husbandry like Oxytetracycline, Lincospectin and Flurefenicole. The presence of resistance to Oxytetracycline, Furazolidone and Lincospectin may indicate the livestock as the source of Salmonella strains for wild animal species in nature or in a zoo environment. There are several documents showing the interactions of infectious agents between farm animals and wildlife. Circulation of such agents within these two sources and distribution of antimicrobial agents between these agents (8, 27, 35, 36) (e.g. feeding animals with infected foods with animal origin) may explain the probable way for the presence of such antimicrobial resistances in zoo species. In the present study, the prevalence of resistance to Nalidixic acid, Streptomycin and Neomycin as the antimicrobials still in use for the treatment of human infections, is another health problem.
These results show the risk of Salmonella strains to transmit these antimicrobial resistance traits to human populations. Also, from another point of view, these results show the possible role of humans as the source of salmonella infections for pet animals, because some investigations have revealed human cases as the source of this infectious agent for animals (37-39). Salmonella serovars may be passed in animal feces and transmitted to humans through fecal-oral contact due to poor hygienic practices, attraction to or curiosity about animals like petting and wild species kept in zoos. Usually, infants and children are at the greatest risk of acquiring zoonotic pathogens from animals (40-42). In many cases, the transmission of zoonotic agents and antimicrobial resistance from non-human species to humans are preventable through improved hygiene and surveillance by strict monitoring programs in petting exhibitions and zoo environment.
CONCLUSION
In conclusion, our results demonstrated a high prevalence of Salmonella serovars Enteritidis & Typhimurium in wild animals living in captivity in Semnan City. Furthermore, resistance to different antibiotics (used in human medicine and veterinary medicine) may represent a different route of infection for the mentioned wild animals. These animals can play an important role in the dissemination and transmission of these resistant serovars into the environment and to other hosts such as livestock and humans via direct/indirect contact.
CONFLICT OF INTEREST STATEMENT
The authors declared that they have no potential conflict of interest with respect to the authorship and/or publication of this article.