Abstract
Anaplasma spp. are widely spread rickettsial bacteria transmitted by ticks and placing high impacts on veterinary and public health. A limited number of studies have been carried out on Anaplasmosis in the central part of Iraq. This study was conducted to determine the presence of Anaplasma spp. in cattle in Al-Qadisiyah province, Iraq. A total of 400 blood specimens were collected from cattle suffering from heavy tick infestation. Cattle were blood-sampled from four hyperendemic areas with ticks. Blood samples were screened using microscopic and polymerase chain reaction (PCR) methods. Diff-quick stained blood smears revealed Anaplasma-like inclusion bodies in 254 (63.5%) samples. According to the 16S rRNA-gene-based PCR analysis, Anaplasma spp. was detected in 124 of the 400 (31%) samples, divided as 96/254 (37.8%) among the microscopical positive samples and 28/146 (19.17%) among the microscopical negative samples. Phylogenetic analysis based on the partial 16S rRNA gene sequencing of ten-PCR positive samples were 99–97% identical to sequences deposited in the GenBank, revealing presence of A. phagocytophilum, A. marginale and unnamed Anaplasma spp. in 40%, 20%, and 40% samples, respectively. Relationships among Anaplasma spp. infections and cattle breed, age, and sex were analyzed. Calves less than one year old showed significantly higher rates (p<0.005) than those from other age groups, whereas sex and breed demonstrated no significant differences (p˃0.001). This study shows that a variety of Anaplasma spp., were endemic in central part of Iraq and is still a hidden problem in cattle in the hyperendemic areas of tick, which requires serious control strategies.
Keywords: Anaplasma phagocytophilum, bovine anaplasmosis, Iraq, phylogenetic analysis
INTRODUCTION
Bovine anaplasmosis is a tick-borne infectious disease caused by five species of the genus Anaplasma (family Anaplasmaceae), including Anaplasma centrale, A. marginale, A. bovis, A. phagocytophilum and A. platys that circulate in cattle worldwide (
1). All species infect blood cells of a variety of domestic and wild animals, as well as humans and show various cell affinity (
2,
3). Bovine anaplasmosis causes significant economic losses through reduction in meat and milk production and high mortality rates in cattle (
4). A. marginale is the main etiological agent of bovine anaplasmosis (formerly known as gall sickness) that infects erythrocytes with a high prevalence reported in many countries (
5). A. phagocytophilum is considered to be a re-emerging host generalist bacterium and is increasingly detected in ticks and variety of hosts, including domestic animals and humans in many parts of the world (
6,
8,
9). In the southern area of Iraq, Hyalomma is a common genus infesting cattle; however, H. anatolicum anatolicum, H. marginatum turanicum, H. asiatcum, H. anatolicum excavatum, Boophilus annulatus and Rhipicephalus turanicus are infesting cattle to a lesser extent (
10,
11). In addition to ticks, the transmission of Anaplasma spp. via bloodsucking flies (Tabanidae family) was also reported (
12). Novel Anaplasma strains related to
A. phagocytophilum were also detected in Hyalomma asiaticum ticks infesting sheep and cattle. However, they are genetically distinct from all other classified and unclassified Anaplasma spp., including the Japanese strains (
13).
Cattle infected with Anaplasma spp. are usually asymptomatic butcanshowmildtomoderateclinical signs including hemolytic anemia, fever, weight loss, decreased milk production, abortion, and prescapular
lymphadenopathies, and the infections may be fatal under other disease complication (
14,
15). Diagnosis of anaplasmosis based on clinical signs and microscopic examinations may not be suitable, if the organism is present in the blood at low levels (
16). Serological tests for diagnosis of anaplasmosis are also hampered by cross-reaction (
17). In Iraq, the occurrence of Anaplasma spp. in domestic ruminants has also been documented (
3,
18,
19). Data concerning the presence, distribution, and molecular typing of Anaplasma spp. is still modest. The aims of this study were to determine the occurrence of A. phagocytophilum and other Anaplasma spp. in cattle of Al-Qadisiyah province, Iraq, to identify the 16S rRNA gene sequence variants and carry out their phylogenetic analysis.
MATERIAL AND METHODS
The survey was performed from April to September (the months of increased tick activity) 2014 to 2016 in mid-Euphrates region of Iraq. The work included the collection of 400 whole blood samples from cattle suffering from tick infestation with variable degrees of anemia-based pale mucus membrane. The samples were obtained from four different locations characterized with tick hyperendemic presence in Al- Qadisiyah province, Iraq. Three milliliters of blood were collected from the jugular vein of each animal using EDTA tubes. Blood smears were prepared by Wedge Method and stained with Diff-quick stains (Syrbio, Switzerland), and the rest of blood amount were stored at -20°C until further processing. DNA extraction was done using a Genomic DNA extraction kit (gSYNC- DNA extraction kit, Geneaid, Seoul, Taiwan) according to the instruction of the manufacturer. The concentration and purity of the extracted DNA were estimated by a NanoDrop (Thermo Fisher, USA). The purified DNA was stored at -20°C, until it was utilized in the molecular analyses.
The DNA of Anaplasma spp. were detected by using the following primers; forward (5'–GGTACC YAC AGA AGA AGT CC-3') and reverse (5'-TAG CAC TCA TCG TTT ACA GC-3'), to amplify a 345 bp fragment of the 16S rRNA gene (
20). The PCR amplification was completed using Multigene thermocycler (Labnet, USA). The reaction mix with a total volume of 50 µl was prepared using 5 µl (100-300 ng) DNA template, 15 pm of each forward and reverse primer in 1.5 µl, 25 µl of 2X Taq master mix with dye, and 17 µl of nuclease free water to amplify the target region under the following conditions: 2 min incubation at 94°C for initial denaturation, followed by 40 cycles of 30 s at 94°C for denaturation, 30 s at 54°C for annealing, and 1 min at 72°C for extension, then finally, the additional extension step for 5 min to complete the DNA extension. The PCR products of 345 bp were analyzed by 1.5% agarose gel electrophoresis.
The amplicons of the positive samples were sent out to Macrogen Company (Seoul, Korea) for DNA sequencing. The phylogenetic analysis was accomplished based on NCBI blast alignment identification (https://blast.ncbi.nlm.nih.gov/Blast.cgi), and the phylogenetic tree was constructed using MEGA 10 software (v10.0.5), following the Maximum likelihood tree (Maximum likelihood)criteria.
For phylogenetic analyses, the newly obtained 10 sequences from cattle were aligned with global Genbank sequences of Anaplasmataceae family from different hosts (bovine, ovine, caprine and canine) and locations. Sequences of the respective genes were aligned in MEGA 10 using the Clustal W program with a default scheme for introducing gaps into the alignment.
Ethical approval for animal care and use was obtained from the College of Veterinary Medicine, University of Al-Qadisiyah.
Statistics analysis
All the result data related to breed, age and sex were analyzed as counts and percentages of PCR positive samples. Chi-square was performed using IBM SPSS software (version 24).
RESULTS
Microscopic screening
Although most cattle were heavily infested with ticks, most infections were subclinical cases but with some mild nonspecific signs. Signs such as pale mucus membranes and weight loss were predominant. Moreover, fever and drop in milk production were also present. A high prevalence of intracellular blood pathogens 254/400 (63.5%), was detected microscopically based on the Diff-quick stained blood smears. Intracytoplasmic Anaplasma-like inclusion bodies were visualized in monocytes, granulocytes, and erythrocytes (
Fig. 1 - A, B, C and D).
Figure 1. A shows intracytoplasmatic inclusion bodies of Anaplasma spp. which are present in monocytes. B and C show Anaplasma spp. in the cytoplasm of neutrophiles from differnt bovine samples (white arrows). D shows Anaplasma spp. in 90% of bovine erythrocytes
Figure 2. Electrophoresis of ethidium bromide stained agarose gel of the 345 bp fragment of the Anaplasma spp. based 16S rRNA gene detected from bovine blood samples in Iraq. Lanes 1–3 and 5–12 are the positive samples. M: the 100 bp molecular marker
Figure 3. Phylogram of Anaplasma spp. based on alignment of 345 bp fragment of the 16S rRNA gene neighboring joining tree was conducted using MEGA 10 Software. The replicate tree percentage in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown above the branches. The evolutionary distances were computed using the maximum-composite-likelihood method. The sequences labeled with red color are the current study isolates
Molecular analysis
The overall prevalence of infection with Anaplasma spp. was 124 out of the 400 (31%) samples according to the PCR results of 16S rRNA gene (
Fig. 2), divided as 96/254 (37.8%) among the microscopical positive samples and 28/146(19.17%) among the microscopical negative samples.
DNA sequencing and phylogenetic analysis
Only ten amplicons obtained by PCR were sent out for sequencing. Three sequence species were identified and matched up with A. phagocytophilum, marginale and unclassified Anaplasma spp. and were 4, 2, and 4 out of 10 samples, respectively. All sequences were deposited in GenBank under the accession numbers shown in
Fig. 3. Comparative analysis of 16S rRNA gene nucleotide sequences was performed with a number of A. phagocytophilum isolates present in the GenBank database, shown in
Table 1. The preliminary results from this study suggest that at least four Anaplasma spp. are circulating in Iraqi cattle population. These species shared a high sequence identity with those of A. phagocytophilum, marginale and undifferentiated Anaplasma spp. of MH155603.1, related to A. platys, A. phagocytophilum and A. boleense (0.05). Anaplasma spp. of MH155604.1 and MH155432.1 have high levels of identity with A. boleense (0.02), and Anaplasma spp. of MH155605.1 has a high percentage of identity with A. marginale and A. centrale (0.01) (
Table 2).
Breed-specific analysis showed that the Anaplasma infection rate increased with age in both breeds of cattle. Considering A. phagocytophilum, A. marginale and Anaplasma spp., infection rate of cattle under one year old was significantly higher (p<0.005) than in the other age groups (
Table 1). Sex and breed showed no significant differences (p<0.001).
Table 1. The percentage of Anaplasma spp. infection in cattle associated with age, sex, and breed of cattle in Al-Qadisiyah province, Iraq
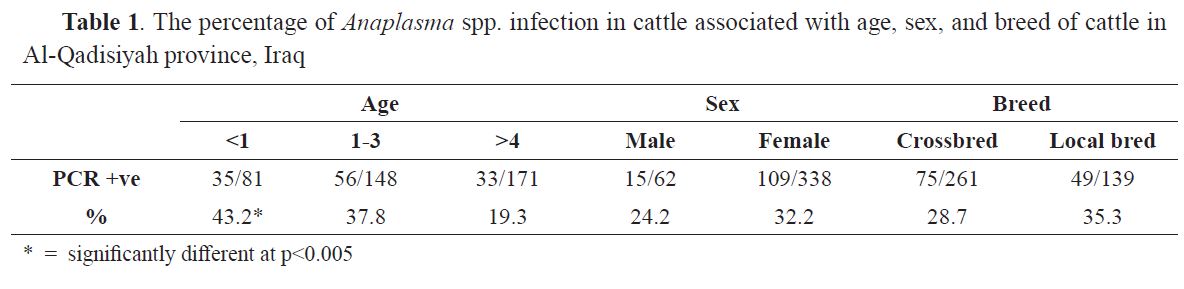
Table 2. Comparison of the sequences obtained from the 16S rRNA partial gene sequencing of Anaplasma genotypes (1-10) with published sequences of Anaplasma spp. in the NCBI GenBank
DISCUSSION
Due to the lack of proper control programs in Iraq, the middle and south of Iraq are increasingly becoming hyperendemic with ticks (
10,
11,
19,
21) that transmit a wide range of veterinary and medically important pathogens (
15,
18,
22). Previous surveys from Iraq reported high infestation rates of cattle with ticks with high infection rates of tick-borne pathogens (
21). Recent studies of A. marginale and A. ovis were detected in significant infection rates in cattle and sheep, respectively (
18,
19). However, the current investigation of bovine anaplasmosis was performed in some fields of Al-Qadisiyah province using microscopic and molecular methods. Clinically, mostexamined cattlewereasymptomatic, but the observed clinical signs agree with (
8,
10,
18) who mentioned that most of natural infections with anaplasmosis pass undetected more often. Molecular methods for detecting Anaplasma spp. in positive and negative microscopic samples provided proof of the realistic occurrence of Anaplasma infection; on the other hand, the microscopic examination of blood smears was not sufficient enough to detect low levels of bacteremia in asymptomatic cattle (
23). Furthermore, blood- cell intracytoplasmic inclusion bodies are often difficult to be differentiated from other hemo- microparasites or blood cell abnormalities (
15). Using the PCR analysis based on universal primers and sequencing proved useful for the identification of uncultured- microorganisms such as Anaplasma. They are also more specific and sensitive for detecting minimum levels of bacteremia in chronic and carrier animals (
20).
However, DNA of Anaplasma spp. was detected in 31% of blood samples based on the molecular assay. This finding agrees with 49%, whereas it is low compared to 77.6% results from Pakistan and Malaysia, respectively (
24–
26). In addition, other researchers recorded 100% of bovine anaplasmosis in some regions (
27). The relatively low infection rate in the current study may be related to uncontrolled using of broad-spectrum antibiotics, such as oxytetracycline in Iraq.
In Iraq, there are no available clinical, pathological, or epidemiological data about A. phagocytophilum infection. Sequencing results show high identities with those of A. marginale (20%), A. phagocytophilum (40%) and Anaplasma spp. (40%), and these results fit well with the observations of (
24,
28,
29) who reported that the prevalence of A. phagocytophilum and A. marginale in cattle blood samples in China were 24.66%, 44%, and 52%, respectively.
The undifferentiated Anaplasma spp. have equal identity values with A. phagocytophilum and A. boleense, but these species are different from those of typical A. phagocytophilum and A. boleense species sequences; these results agree with those reported in Japan (
13). Only 2% of Anaplasma spp. sequences were closely related to A. centrale (0.01 distance) (
Table 2). This finding is confirmed by the study of Jassem and Agaar (
27), who failed to find A. centrale during their investigation and suggested that the bacterial levels were low in the blood samples in cattle from the middle region of Iraq.
Based on the statistical results, different breeds of cattle may show varying levels of resistance against anaplasmosis. Native breeds of cattle have been reported to show lower resistance to infection than crossbred cattle. High variety of infection susceptibility has been reported in many other studies (
27). The results of the gender-wise prevalence revealed that females were more susceptible to the infection than males, but with no significant differences. These results are in agreement with Tay et al. (
2). who published that there are no significant differences between genders. In general, the results of the phylogenetic analysis suggested that at least four Anaplasma species circulated in our cattle. A. phagocytophilum was a frequently isolated organism from the sampled cattle. This study describes, for the first time, the presence of A. phagocytophilum in Iraq suggesting a wider area of distribution of this pathogen than previously assumed.
CONCLUSION
The study findings suggest that at least four Anaplasma species are circulating in cattle from Iraq, mainly A. phagocytophilum, A. marginale, and other Anaplasma spp. The detection of A. phagocytophilum in the cattle of the present study represents the first-time identification study in Iraq. Anaplasma spp. were detected displaying high sequence similarity to those of A. platys, A. phagocytophilum, A. marginale, A. centrale and A. ovis. Further investigations should be conducted concerning the zoonotic potential of the A. phagocytophilum agent detected in this study and the real role of cattle and cattle products in the epidemiology of human granulocytic anaplasmosis in Iraq. Further studies on identification and characterization of Anaplasma spp. and the essential vector ticks are necessary to be performed for the purpose of finding novel control and eradication programs against this parasite.
CONFLICT OF INTEREST
The authors declared that they have no potential conflict of interest with respect to the authorship and/or publication of this article.
ACKNOWLEDGEMENT
We thank Moneer Abd-Alameer, (Head of Department of Parasitology, College of Veterinary Medicine, University of Al-Qadisiyah) for allowing the senior authors to work in their laboratory. All lab. carers, who supported us with material, are acknowledged with thanks. Our thanks also go to animals owners for allowing examining and sampling of their animals.

 10.2478/macvetrev-2019-0023
10.2478/macvetrev-2019-0023
