Abstract
For the first time in Iraq, we identified in March, 2018 the presence of a highly virulent avian influenza virus (AIV), H5N1 (Clade 2.3.2.1c), causing highly pathogenic avian influenza (HPAI) in poultry farms, Iraq, . The identification of the virus was done using a rapid serological test, a real time-qPCR, and glycoprotein gene sequencing. Using sequencing and phylogenetic analyses, the clade 2.3.2.1c virus was recorded to be clustered, with high similarity to Asian and West African AIV, HPAI H5N1 from Ivory Coast identified in 2015. According to our knowledge, there was no previous detection of the clade 2.3.2.1c made in Iraq. Our results provide evidence that high risk of HPAI H5 outbreaks might be present in Iraq, and this needs to lead to high quality surveillance targeting of wild and domestic birds for early diagnosis of HPAI. The current work provides feasible and accurate approaches for understanding the evolution of HPAI H5 virus in different countries around the world.
Keywords: avian influenza, H5N1, phylogeny
INTRODUCTION
The Orthomyxoviridae family consists of influenza A viruses that have single-stranded negative-sense RNA, with eight genomic-based segments. These viruses are arranged into 18 HA (H1 to H18) and 11 NA (N1 to N11) subtypes detected using haemagglutinin (HA) and neuraminidase (NA) envelope glycoproteins classification on the basis of phylogenetic and antigenic characteristics (
1). All of these AIV subtypes are stored in wide ranges of aquatic birds, acting as reservoirs. During 2-AIV-genotype-based infection affecting a single cell or aquatic birds in the wild, genetic reassortment or directional selection pressures are performed on the genetic materials of these viruses. This leads to induction of a certain genetic evolution, producing a virus that can escape immunity by substitutions of few amino acids (
2).
Moving of animals was considered an illegal act as those animals may maintain infections by this virus. Using HA-gene-based sequencing, the Lebanese H5N1 HPAI virus was detected to be from clade 2.3.2.1c, which was highly similar to the Bulgaria, Romania, and Turkey viruses identified in 2015 from samples collected from wild and domestic birds (
15). In 2015 and 2016, the countries of Cambodia, China, Vietnam, and West Africa were affected by the same clade of the H5N1 HPAI virus. Since that time, Lebanon has declared containment of the outbreaks (
16). Establishment and spread of H5N1 HPAI virus in the region is maintained via the virus incursions (
12).
Correct diagnosis of the virus is made using HA subtype detection. HPAI-phenotype-based detection of H5 and H7 has been performed in free natural environment only (
3). The continuous evolution of the gs/GD lineage has been determined to result into different clades, such as (2.2.1.2, 2.3.2.1, and 2.3.4.4) that are considered as major clades and (1.1.2, 2.1.3.2, and 7.2) that are the minor ones. Those clades cause endemic AI in Asia, Africa, and Middle East (
4). The infection and the spread of the virus may be induced by infected- bird-spillover transmission and migration of wild birds, leading to wide areas of transmission of these viruses (
4).
Some of these viruses are classified as low pathogenic AI (LPAI), showing less severe clinical signs than HPAI. The H5 and H7 subtypes are the common-known HPAI viruses spread around the world (
5). The H5 subtype was recently a subject of interest because of the potential for transmission in humans, as evidenced by the 1997 Hong Kong incident in which 6 of 18 human cases died (
6). Emergence and reemergence of the HPAI virus is a concern for countries and governments, because of its effect on the health of people and the economy. Growth of the domestic poultry sector in response to the rapid increase in global demand for protein, as well as climate change, the increase of international trade and land cover fragmentation, have been considered to be important risk factors for the continuous global increase in the incidence of emerging zoonoses such as avian influenza (AI) viruses, including Middle East (ME) countries (
7). The first novel emergence of H5N1 HPAIV occurred in commercial domesticated geese in the Guandong province in China, in 1996 (
8). Since then, the ancestral strain of H5N1 HPAIV has continued to circulate within Asia and consequently has passed through wide ranges of genetic changes leading to the evolution of various virus lineages, sometimes also referred to as clades (
9) AI surveillance activities included active and passive strategies, with few countries making use of molecular techniques, whereas most made use of serologic tests for detection (
10). Within the ME, Turkey was the first country to report H5N1 HPAIV in backyard poultry, in October 2005. Subsequently, more than 1000 cases were recorded in Egypt, Israel, Iran, Iraq, Kuwait, Jordan, Sudan, the Palestinian Territories and Djibouti during November, 2005 and April, 2006. In 2007, Saudi Arabia was the last country to report a series of H5N1 outbreaks, whereas Libya reported their first single introduction in 2014 (
11).
In countries having outbreaks and endemic problems of AIV, especially those neighboring Iraq, the evolution based on the genetic and antigenic characteristics of H5N1 viruses has been thoroughly recorded. However, Iraq has limited information about this evolution occurring with egards to the H5N1 viruses. According to these criteria, gene-based sequencing of the H5N1 clade 2.3.2.1c was performed. The current research is aimed at drawing a complete picture of the serotypes that infect poultry in Iraq, for the purpose of preparing a good strategy for eliminating of this virus and developing guidance for the control and the prevention of the H5N1 viruses.
MATERIAL AND METHODS
Poultry farms
The current work was done in accordance with the ethics guidelines of national and global regulations for animal care and use. An outbreak occurred during March 2018, in Baghdad, the capital of Iraq, that consisted of 4 caged houses for 18 weeks layers with a total number of 80,000 hens. The symptoms recorded were respiratory symptoms including birds with dyspnea, recumbency, head tremors, decreased awareness, and somnolence. Additionally, beaks and claws were removed from four freshly dead birds, then the birds were sealed in a plastic bag, removed from the barn, disinfected, sealed in a second bag, and again disinfected. The company placed the farm under temporary quarantine pending test results. The morbidity and mortality rate were calculated. All farms within a 10-km control area were quarantined, and infected farms were depopulated.
Serological diagnosis
Sixty four cloacal swabs were used for serological diagnosis of the AI virus antigen using Rapid H5 Ag Test Kit (Elabscience, USA). If the kit window showed two color bands that meant a positive result, while the presence of only one color band in front of C letter meant a negative result.
RNA extraction
The samples (brains, lungs, tracheas and rectums) were pooled on FTA spots (Whatman® FTA® card technology). Four samples were pooled for each area. The compositions of the FTA cards include some chemicals that act on lysing the cells, denaturing proteins, and protecting nucleic acids against the activities of nucleases, the oxidizing and UV-based damaging effects. One of the benefits of FTA cards is that they deactivate sample-related microorganisms rapidly, such as blood-borne pathogens. Another benefit is that nucleic acids are entrapped in the matrix fibers preventing transport-based damages, allowing immediate processing, or enhancing long- term ambient storage. Four cards were shipped to AniCon Labor GmbH I Muehlenstraße 13a I 49685 Hoeltinghausen I Germany, then viral RNA was extracted with the manufacturer’s instructions of Kylt® RNA / DNA Purification kit.
Real-time PCR
The real-time PCR reactions were run by AniCon Labor GmbH, Germany. To sumarize, species-specific and variant-specific Real-Time RT-PCR method was used to detect Influenza Virus type A, specific Real- Time RT-PCR Hybridization probe-based chemistry was used with the following primers: (Kylt® Influenza A - H5 / N1).
RESULTS
Clinical signs and postmortem exam
The disaster started with infection of about 80,000 hens, when the flock suffered from signs of avian influenza and the mortality rate reached 90% for about 10 days, showing the following symptoms: decreased feed consumption, severe rales, sinusitis, edema of the head and wattles, diarrhea and sudden death. The postmortem findings included sinusitis with mucopurulent to caseous exudate, fibrinopurulent pericarditis, congestive, hemorrhagic and necrotic changes on the skin and the intestinal tract, hemorrhages in proventriculus and heart.
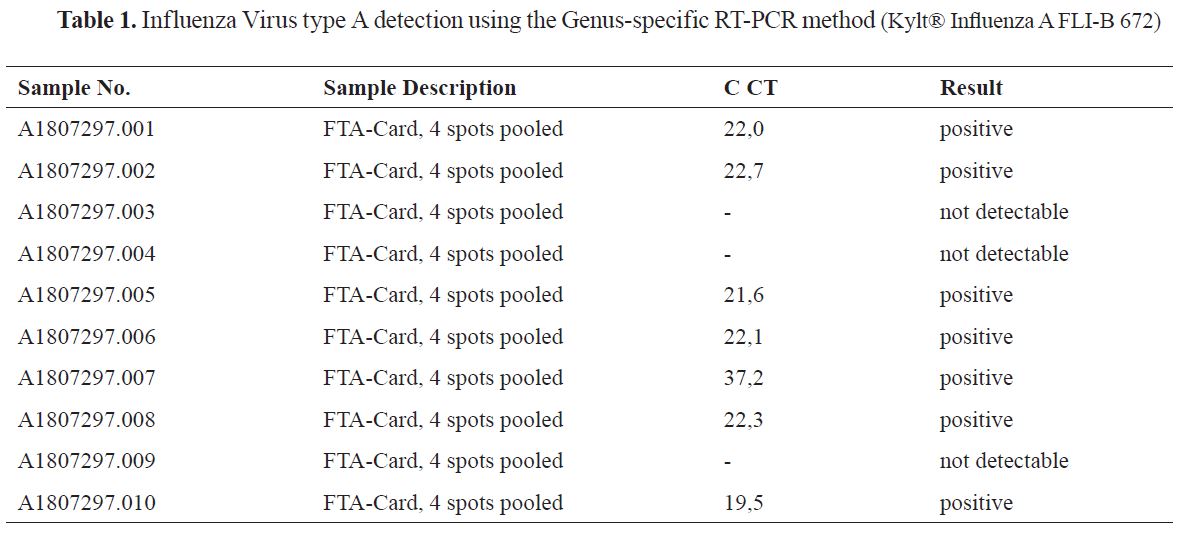
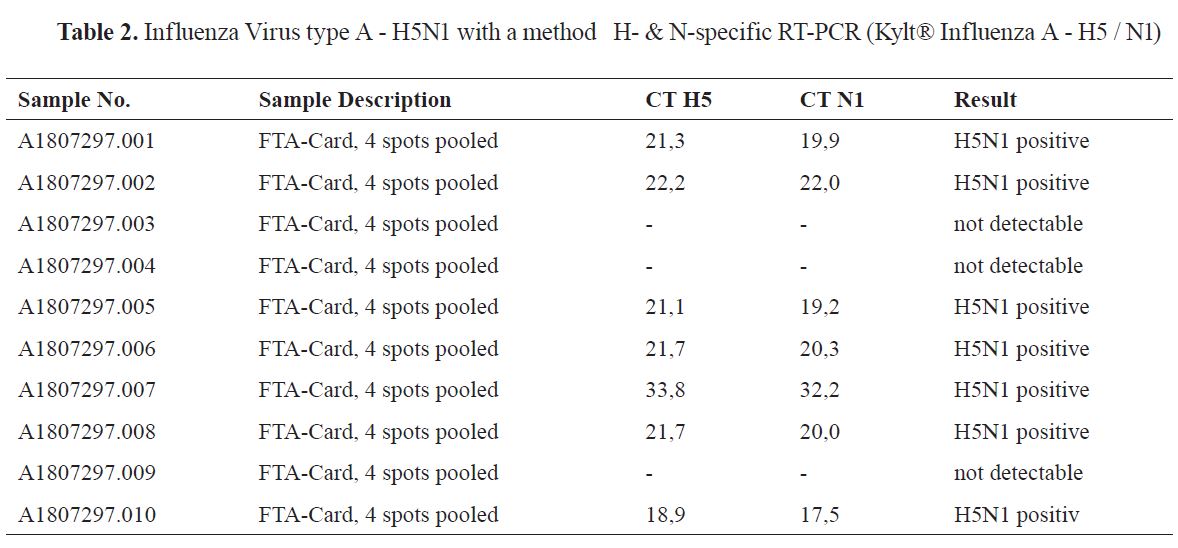
Figure 1. Phylogenetic tree that is based on the amplified nucleotide sequence coding for the hemagglutinin (HA) glycoprotein of Influenza A Virus (type H5), using MEGA v10 software, the RNA extracted from our sample (A1807297.001) belongs to HPAI H5 Clade 2.3.2.1c
Real-time PCR
The viral RNA from the FTA card (4 spots pooled) with the samples number (1-10) showed positive results for seven samples out of ten, using the Genus-specific Real-Time RT-PCR (Kylt® Influenza A FLI- B 672) method as shown in the
Table 1.
Also, all the samples (1-10) were tested for: Influenza Virus type A - H5N1 with a method H- & N- specific Real-Time RT-PCR (Kylt® Influenza A - H5 / N1), with seven of them being positive results as in
Table 2.
Sequencing of HA gene products and phylogenetic analysis
A 900 bp fragment of the HA gene of Influenza A type H5 has been sequenced and was phylogenetically analyzed based on the comparison to known reference strains. Based on the amplified nucleotide sequence coding for the hemagglutinin (HA) glycoprotein of Influenza A Virus (type H5), the RNA extracted from our sample belongs to HPAI H5 Clade 2.3.2.1c and is most related to strains that had been isolated from peafowl in the Kirkuk region in Iraq, as shown in
Fig. 1.
Gene bank accession number
The hemagglutinin (HA) glycoprotein of Influenza A Virus (type H5) were submitted to the NCBI geneBank data base. The GenBank accession number was MK034857.1 under the name Influenza A virus (A /chicken /Baghdad/ Bag-1/2018(H5N1)
DISCUSSION
For the last 10 years AIV has been recorded which has lead to major economic crisis in poultry production lines in different Iraqi provinces, but The H5N1 has been recorded in 2015- 2016 only with 11 outbreaks.
Observed mortality rates ranged from 10% to 100% (
12). Genetic-based characterization of the Iraqi AIV has not been done to date. According to symptoms, gross lesion, high morbidity and mortality rate (90%), the suspected diagnosis was that the infection is avian influenza. However, unfortunately, we don’t have all the facilities in Iraq to make accurate diagnosis. So, the aim of this study was detection and genetic characterization of H5N1 after preliminary determination of infection with a rapid H5N1 kit.
The current work gives genetic information of the H5N1 virus circulating in poultry farms. We precisely detected, in Iraq, the presence of the H5N1 Clade 2.3.2.1 c in cooperation with AniCon Labor GmbH, Germany. This study characterizes the clinical signs, gross lesions, and antigen distribution that were detected in a commercial hens flock, after infection with H5N1 HPAI. After the initial positive diagnosis of HPAI, the inspection of poultry farms in a surrounding 10 km zone identified additional, previously unrecognized flocks infected with H5N1.
The available information revealed that all countries surrounding Iraq were infected with many kinds and multiple times with avian influenza. The H5N1 HPAI virus is known to be endemic in some regions of South and Southeast Asia, which commonly applies to clades 2.1.1, 2.3.1 and 2.3.2.1a and c; however, Egypt is known to be endemic with clades 2.2.1 and 2.2.1.2, but West Africa show infections with clade2.3.2.1c (
12). Europe (Bulgaria, Romania and Turkey) has been invaded with clade 2.3.2.1c affecting wild birds and poultry (
13). The Middle East has been infected with the virus since 2006, affecting poultry and wild birds reported officially or unofficially. Observation of H5N1 HPAI invasions have been noticed since 2006 in the Gaza Strip, the West Bank, Jordan, Israel, Iran, Kuwait, Turkey, Saudi Arabia and Yemen. However, the infection by H5N1 HPAI in Syria has not officially been declared. In 2015, H5N1 HPAI viruses closely matching to those identified in Egypt, clade 2.2.1, were isolated from outbreaks recorded in the West Bank, Israel, and the Gaza Strip. Turkey and Iran, in 2015, had outbreaks of the H5N1 HPAI. H5N1 HPAI outbreaks were recorded in the southwest of France during October, 2015 and April, 2016; however, the viruses detected according to genetic bases were from the European strains, but not from the ones mentioned (
14). Affecting many poultry farms, the HPAI virus was recorded for the first time in Lebanon in 2016.
CONCLUSION
Our results showed that the virus detected in the current study samples belongs to the clade 2.3.2.1c of the HPAI H5N1. The highest similarity of our strain was noticed with sequences identified in peafowl in the Kirkuk region in Iraq. Although this study provides new information about the pathology and antigen distribution of this novel virus, additional work is likely needed to characterize this pathogen.
CONFLICT OF INTEREST
The authors declared that they have no potential conflict of interest with respect to the authorship and/or publication of this article.
ACKNOWLEDGEMENTS
The authors thank the departmental Lab of Poultry Diseases, College of Veterinary Medicine, University of Al- Qadisiyah, Diwaniyah, Iraq.

 10.2478/macvetrev-2019-0024
10.2478/macvetrev-2019-0024

