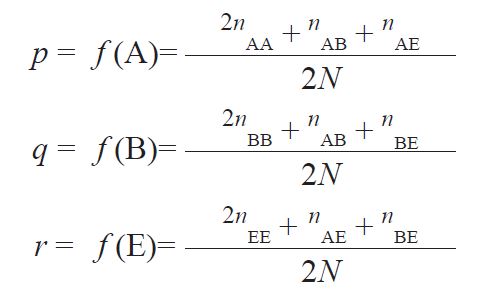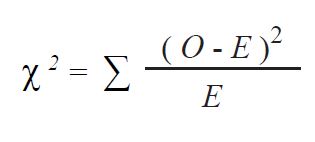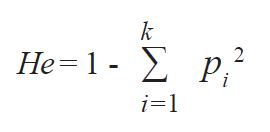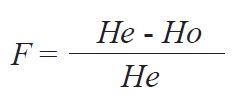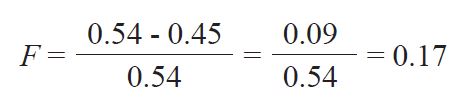Abstract
The bovine kappa-casein (κ-CN) is a phospho-protein with 169 amino acids encoded by the CSN3 gene. The two most common gene variants in the HF breed are CSN3*A and CSN3*B while CSN3*E has been found with lower frequency. The aim of this study was to optimize a laboratory method for genotyping of these three alleles as well as to determine their genotype and allele frequencies in the HF cattle population in the Republic of North Macedonia. Genomic DNA was extracted from full blood from 250 cows. The target DNA sequence was amplified with newly designed pair of primers and the products were subjected to enzymatic restriction with HindIII and HaeIII endonucleases. Genotype determination was achieved in all animals. The primers successfully amplified a fragment of 458 bp and the digestion of this fragment with both endonucleases enabled differentiation of five different genotypes with the following observed frequencies: AA (0.39), AB (0.29), BB (0.16), AE (0.10), and BE (0.06). The estimated allele frequencies were: CSN3*A (0.584), CSN3*B (0.336) and CSN3*E (0.08). The observed genotype frequencies differed significantly (P<0.01) from those that would be expected under HW equilibrium, while the fixation index (F=0.17) indicated moderate heterozygosity deficiency. Nevertheless, the CSN3*B allele was present with relatively high frequency which should be used to positively select for its carriers, since increasing its frequency could help to improve the rheological properties of the milk intended for cheese production.
Keywords: CSN3, genetic polymorphism, Holstein-Friesian cattle, kappa-casein, milk protein
INTRODUCTION
The most important milk proteins are caseins which are produced by the mammary gland secretory cells. They constitute about 80% of the bovine milk proteins (
1) and are divided into four main fractions: αs2 -CN, β-CN, αs2 -CN, and κ-CN.
The kappa-casein (κ-CN) fraction which constitutes around 12% of the total caseins found in the bovine milk (
2) is a phospho-protein with 169 amino acid residues (
3) located predominantly on casein micelle surface and is specific substrate of the chymosin which hydrolyses its amino acid chain at position Phe105- Met106 and yields insoluble para-κ-CN (amino acid 1-105) and soluble caseino-macropeptide-CMP (amino acid 106-169) (
4,
5).
The bovine κ-CN is encoded by the
CSN3 gene located on BTA6 (
6,
7,
8,
9). This gene is around 13 kb in length and is divided in transcription unit (5 exons and 4 introns) and 5’ and 3’ untranslated regions (
10). The fourth exon which is 517 bp long (
11) harbors all the 11 non-synonymous single- nucleotide substitutions which code 11 different variants of the mature κ-CN protein identified so far in the
Bos genus (3): А, B, C, E, F1, F2, G1, G2, H, I and J. In a more recent review on milk protein polymorphism in cattle (
12) it has been suggested two more alleles to be included to this list since they are non-synonymous mutations in this exon namely:
CSN3*B2 and
CSN3*D. In addition, one more synonymous nucleotide substitution has been identified, namely
CSN3*A1 of Damiani et al. (
13) or
CSN3*A1 of Prinzenberg et al. (
14) which does not modify the correspondent amino acid.
The two most common protein variants in the HF breed are κ-CN A and κ-CN B (3, 14) while κ-CN E has been found with lower frequency in this breed (
12). With the exception of the Jersey breed the variant κ- CN A is the most common among dairy cattle breeds (
16,
17,
18). The κ-CN B protein variant differs from κ-CN A at two amino acid positions: The136 is substituted with Ile and Asp148 is exchanged with Ala. The κ-CN E variant differs from κ-CN A at amino acid position 155 where Ser is substituted with Gly (
12). The
CSN3*B allele is used as a genetic marker in dairy cattle breeding programs because the milk with κ- CN B protein variant has been shown to have better rheological properties such as shorter rennet coagulation time and higher yield during cheese production (
1,
19, 2
#ref10) when compared to milk with κ-CN A variant. Bovenhuis et al. (
21) suggested that the favourable milk protein genotype κ-CN BB should be included in the criteria for selection of dairy cattle because of economic interest.
The aim of this study was to optimize laboratory method for genotyping of the most common κ-CN variants in the HF cattle population in the Republic of North Macedonia as well as to determine the genotype and allele frequency at this locus. We focused on the CSN3*A, B and E alleles because they have been reported in the literature as the most frequent variants in different dairy cattle populations.
MATERIAL AND METHODS
DNA extraction and quantification
Genomic DNA was extracted from blood obtained by venipuncture of the jugular or the coccygeal vein from 250 cows, selected randomly from five cattle farms in the Republic of North Macedonia. The blood was drawn in vacutainers with anticoagulant (EDTA) and was stored at +4°C until extraction. The DNA was extracted from blood using two different methods: i) Phenol-Clorophorm- Isoamil alcohol followed by ethanol precipitation, and ii) with commercial DNA extraction kit. The amount and the purity of the extracted DNA was determined with spectrophotometer.
PCR amplification of the CSN3 locus
The primes used for amplification (KCN-F: GGTCACCTGCCCAAATTCTTCAA and KCN-R: AGCCCATTTCGCCTTCTCTGT) were designed using the Primer Premier software (Premier Biosoft International) based on GenBank sequence X14908.1. (
10). The coefficients of hairpin formation, self- dimerization and creation of cross-dimers as well as the primer’s optimal annealing temperature necessary to design the reaction conditions for the thermo-cycling protocol, were determined with the same software. These primers amplified a region of 458 bp of the 4th exon of the bovine
CSN3 gene. Part of this nucleotide sequence with the primer annealing positions and the restriction endonucleases cleavage sites are shown in
Fig. 1.

Figure 1. Part of the nucleotide sequence GenBank X14908.1 that corresponds to CSN3*A allele. The primer binding sites are denoted with boxes, the restriction sites of the enzymes are marked with arrows and the positions of the two nucleotide substitutes that create additional restriction sites for alleles CSN3*B or CSN3*E are shown with bold underlined letters
The amplifications were2 prepared in total volume of 20 µl containing 1 X PCR buffer, 200 µM dNTP, 2.0 mM MgCl , 0.6 U DNA Polymerase, 0.2 µM of each oligonucleotide and 40-50 ng genomic DNA. The following thermal protocol was applied: initial denaturation of 95°C/5 min. then the
Taq DNA Polymerase was added followed by 35 cycles of 94°C/45 sec., 56°C/45 sec., 72°C/1 min., and final elongation step of 72°C/5 min. on Biometra TPersonal Thermocycler (Biometra GmbH, Germany). The amplified DNA fragments were checked by staining with ethidium bromide on % (w/v) agar-gel followed by visualization on a UV transiluminator (
Figure 2). A 100 bp DNA ladder was lined up as molecular size marker.
Genotyping of the amplified products
In order to detect the three alleles and their combinations of genotypes, Restriction Fragment Length Polymorphism (RFLP) analysis was carried out with two different restriction endonucleases. Initially, each PCR product was digested with the
HindIII enzyme (Thermo Scientific) which enables distinction of
CSN3*A or
CSN3*E allele carriers from those that are carriers of the
CSN3*B allele. This enzyme did not enable distinction of the
CSN3*A from
CSN3*E allele since it has the same cleavage site in both alleles and consequently it yields same restriction fragment lengths from both alleles. xConsequently, those samples where the variant
CSN3*A was detected with this enzyme (genotypes classified as AA and AB), were further digested with the
HaeIII enzyme in a separate reaction in order to enable distinction of
CSN3*A from
CSN3*E variant (since the
CSN3*E allele has two cleavage sites for this enzyme and yields three fragments, while the
CSN3*A variant has only one cleavage site and yields two fragments) as shown in
Fig. 1.
In this study GenBank sequence X14908.1 was used as a reference sequence to design the primers and to predict the restriction patterns. In this sequence the following nucleotide positions were used to differentiate the three CSN3 alleles:
-
the primers amplified the region between the nucleotide positions 5105 and 5562;
-
between nucleotides 5221 and 5222 there is a cleavage site for HaeIII (GG/CC) in all three studied
CSN3 alleles;
-
at nucleotide position 5345 the transversion A→C in the CSN3*B allele (Asp148Ala) creates cleavage site for HindIII (A/AGCTT) while the other two alleles remain undigested at this position;
-
at nucleotide position 5365 the transition A→G in the CSN3*E allele (Ser155Gly) creates additional cleavage site for HaeIII (GG/CC) while the other two alleles remain undigested at this position.
For each genotype, the expected fragment sizes after digestion with both enzymes are shown in
Table 1.
Table 1. Expected fragment sizes (in bp) corresponding to different CSN3 genotypes after digestion of a 458 bp PCR product with two restriction enzymes
The digestion reactions were prepared in total volume of 20 µl containing 2 µl 10X Buffer, 8 µl PCR product, 1-2 µl restriction enzyme, and 8-9 µl ddH O. The incubations were carried out at 37°C for a period of 3 h. Digested products were analysed using electrophoresis on 2.5% agarose gel stained with ethidium bromide. Band patterns were visualized via UV transilluminator photo documentation system (
Fig. 3).
Statistical analysis
The observed number of animals for each of the five detected genotypes was calculated by direct counting. The frequencies of each of the three alleles were estimated by allele counting method (
22) by adding twice the number of homozygotes to the number of heterozygotes that possess the allele and divide this sum by twice the number of animals in the sample or:
where:
n is the number of animals possessing the genotype, and N is the number of animals in the sample.
These estimated allele frequencies p, q, and r were used to calculate the expected number of animals for each genotype as follows: AA = N x p2, AB = N x 2pq, BB = N x q2, AE = N x 2pr, BE = N x 2qr and EE = N x r2.
The probability of Hardy-Weinberg equilibrium associated with the observed genotype frequencies was calculated by the Chi-squared (χ2) goodness- of-fit test (
23) as follows:
where:
χ 2 : Chi-squared test statistic
O: Observed number of genotypes
E: Expected number of genotypes
The χ2 test statistic had k - 1 - m degrees of freedom, where k is the number of genotypes and m is the number of independent allele frequencies estimated from the data (
24).
To determine the level of departure from the HW expectations in the studied population, the average expected heterozygosity (He) or Nei’s gene diversity was calculated by adding up the expected frequencies of each possible homozygous genotype and subtracting this sum from one (
23) o
where:
k is thei number of alleles at the locus, p 2 is the expected genotype frequency of homozygotes based on allele frequencies, and ∑ 𝑘𝑘 𝑖𝑖=1 indicates summation of the frequencies of the k homozygous genotypes.
The observed heterozygosity (Ho) was calculated by adding the frequencies of the three observed heterozygous genotypes or Ho = f(AB) + f(AE) + f(BE) (
23). From these data the fixation index - F was calculated as follows:
where:
He is the H-W expected frequency of heterozygotes based on estimated allele frequencies and Ho is the observed frequency of heterozygotes.
RESULTS
The primers KCN-F and KCN-R that were designed in this study successfully amplified a fragment of 458 bp in length as illustrated in
Fig. 2.
Figure 2. Representative agarose gel showing PCR amplification of 458 bp fragment of the bovine CSN3 gene. Lanes 1-4 and 6-8: 458 bp fragment, lane 5: GeneRuler 100 bp DNA ladder (Thermo Scientific)
Figure 3. Digestion patterns of exon 4 of the bovine CSN3 gene. Lanes 1, 4, 5 and 11- genotype AA; lanes 3 and 10 - genotype AB, lane 7 - genotype BB; lanes 2 and 8 - genotype AE; lane 9 - genotype BE; lane 6 - GeneRuler 100 bp DNA ladder (Thermo Scientific). The upper half of the agarose gel represents digestion with HindIII and on the lower half the same samples in the same order are digested with HaeIII.
The PCR products were further digested with
HindIII and
HaeIII enzymes. Considering the information of both digestions in terms of number and sizes of the obtained fragments (
Table 1), it was straightforward to identify 5 different genotypes as shown in
Fig. 3. The genotype EE was not detected in the studied population.
Observed genotype counts and frequencies as well as estimated allele frequencies are shown in
Table 2.
From the estimated allele frequencies, the number of animals which under HWE would be expected for each genotype was calculated as shown in
Table 3.
Since the critical value of χ2 0.01,3 = 11.345 it can be concluded that the H-W expected genotype frequencies are not present in the studied population. The expected (He) and the observed ( Ho) heterozygosity were calculated as follows:
He = 1 - (0.341 + 0.113 + 0.006) = 0.54, and
Ho = 0.29 + 0.1 + 0.06 = 0.45
The fixation index (F) was calculated as:
This value indicates moderate (17%) heterozygosity deficiency relative to HW expectations.
DISCUSSION
One of the major effects of the milk protein polymorphism on cattle traits with economic interest is their influence on milk renneting capability and yield during cheese production. The κ-CN fraction, located mostly on casein micelle surface is the specific substrate of the chymosin, the hydrolytic enzyme that has the crucial role in initial phase of the cheese production - the rennet formation (
25).
It has been reported that milk with
CSN3*BB genotype had significantly higher casein content (
26) and better milk rennet coagulation properties in terms of shorter rennet clotting time, higher curd firmness and higher cheese yield (
20-
32). These differences are related to the micelle size and the glycosylation degree of the coded protein (
33).
The primers that were designed in this study successfully amplified a fragment of 458 bp from the fourth exon of the bovine CSN3 gene that included the nucleotide substitutions that differentiate the three investigated alleles. Genotype determination was achieved in all animals of the investigated population. For RFLP genotyping of the six genotypes, it was necessary each PCR product to be digested with the HindIII enzyme which enabled distinction of CSN3*A and CSN3*B alleles, but could not make distinction between alleles CSN3*A and CSN3*Е. For that purpose, it was necessary to further digest those samples carrying the allele CSN*3 A (genotypes AA and AB) with HaeIII which has one cleavage site for the alleles CSN*3 A and CSN*3 B and two cleavage sites for allele CSN*3 E.
Similar approaches for CSN3 genotyping have also been used previously with different restriction enzymes (1, 34-43).
In this study, the
CSN3*A allele was found to be more commonly distributed (0.584) than the
CSN3*B allele (0.336), while the
CSN3*E allele was observed with lowest frequency (0.08). These results are in accordance with the previously published studies in which in HF cattle population in different countries the
CSN3*A allele (genotypes AA and AB) have been more frequently observed than the
CSN3*B allele (genotype BB) as summarized in
Table 4.
With the exception of the Jersey cattle (
26,
45,
52) and the Brown Swiss cattle (
26) in which the
CSN3*B allele has been reported to be more common, the
CSN3*A variant tends to be predominant in most dairy breeds (
16-
18).
Moreover, some less-common
CSN3 alleles might affect milk rheological properties. For instance, Erhardt et al. (
53) reported that in the Pinzgauer breed the
CSN3*G allele had negative effect on milk coagulation properties. Similarly, Caroli et al. (
54) and Jensen et al. (
25) detected a negative effect of
CSN3*E on milk coagulation properties in the Italian and Danish Holstein- Friesian populations, respectively.
It is worth noting that the majority of the previously published studies are concerned about discriminating
CSN3*A and
CSN3*B alleles in different cattle populations, while only a few of them deal with detection of other alleles of the
CSN3 locus such as Pacheco Contreras et al. (
2), Barroso et al. (
55).
The frequency for the
CSN3*E allele of 0.08 found in this study is identical with the results for this allele reported by Jann et al. (
56) and lower than Boetcher et al. (
57) who reported a frequency of 0.32. Soria et al. (
43), in the Argentinian Holstein population reported a frequency of 50 % AA, 40 % AB and 10% AE κ-CN genotypes.
In our opinion, besides detecting variants
CSN3*A and
B, it is important to genotype at least for those alleles of this locus that were reported to have negative effects on some milk properties in different cattle populations (
25,
53,
54). Furthermore, it would also be more informative approach, whenever possible, to use direct sequencing of the
CSN3 gene such as Schlieben et al. (
58) or Chen et al. (
59) since this method enables discovery of new nucleotide variations while PCR- RFLP analysis is limited only to those that have already been reported.
Although κ-CN is the most important factor in the renneting process, interactions with other milk protein variants have to be considered. For instance, Comin et al. (
60) reported that
CSN3 and
CSN2 are strongly associated with milk coagulation traits and milk and protein yields, respectively, and concluded that for coagulation time and curd firmness, the best composite genotypes were those with at least one B allele at both loci. In addition, because positive correlations have also been demonstrated between β-LG BB genotype and higher cheese yield and casein number (reviewed by Buchberger and Dovc;
20) these authors conclude that κ-CN B and β-LG B are the most advantageous variants with respect to milk’s cheese making ability, and they propose that due to the tight linkage that exists between the casein loci, a more extensive study of their haplotype effects is needed.
In this study a departure from the HW equilibrium and moderate heterozygosity deficiency was observed for the investigated genotypes of the CSN3 locus. This could be due to genetic drift as a result of finite population size, population subdivision or due to non-random mating or inbreeding.
CONCLUSION
The primers designed in this study successfully amplified a 458 bp fragment of the fourth exon of the bovine CSN3 gene which harbours the nucleotide variations among the three CSN3 alleles A, B, and E. In the studied population five out of six possible genotypes were identified and departure from the HW equilibrium was observed. Also, a moderate heterozygote deficiency was detected. The allele CSN3*A was the most commonly distributed followed by the CSN3*B while the CSN3*E allele was observed with the lowest frequency. Nevertheless, the CSN3*B allele was present with relatively high frequency which should be used to positively select for its carrier animals, since increasing its frequency could help to improve the rheological properties of the milk intended for cheese production.
CONFLICT OF INTEREST
The authors declared that they have no potential conflict of interest with respect to the authorship and/or publication of this article.
ACKNOWLEDGEMENTS
The authors would like to acknowledge and express their gratitude to the Faculty of Veterinary Medicine in Skopje for providing the financial grant for this research, as well as the national Holstein-Friesian cattle breeders for their support in collecting the blood samples.

 10.2478/macvetrev-2020-0013
10.2478/macvetrev-2020-0013