This paper presents an outbreak provoked by methicillin-resistant strains of coagulase-positive S. aureus (CPS), produced staphylococcal enterotoxins (se) in pig meatballs and potato salad consumed from 70 people in the village Mamarchevo, Bulgaria. Eighteen women aged 50 to 70 years, and two children aged 4 and 5 years have demonstrated a severe malaise with vomiting and indigestion. Two food samples and isolates of CPS were received in the laboratory of Bulgarian Food Safety Agency. Both samples were found to have a high level of CPS. The level of S. aureus contamination in the potato salad was 8.3 logs CFU/g and 7.7 logs CFU/g in roasted meatballs, which was a significant reason to doubt the production of the toxin. The samples were analyzed according to the European Screening Method v5 using mini VIDAS SET2. The results showed a presence of staphylococcal enterotoxin (TV 2.67 for meatballs and TV 3.27 for potato salad), which was the reason for the ensuing food intoxication. EURL CPS applying quantitative indirect sandwich-type ELISA confirmed the presence of sea, sec and sed in the potato salad and sea and sed in the roasted meatballs. Two CPS isolates were confirmed as S. aureus by a species-specific 23S rRNA targeted PCR test. Real-time PCR method detected sea, sed, seg, sei, sej, and ser genes in S. aureus strains, found in both matrixes. Multiplex PCR method proved the existence of the mecA gene in both S. aureus strains. Resistance to cefoxitin (>16 mg/L), penicillin (>2 mg/L), kanamycin (64 mg/L) and sulfamethoxazole (>512 mg/L) was found.
Staphylococcal intoxication is one of the most common food poisoning, dating back to 1914 when Dr. M.A. Barber first associated this poisoning with the production of
Staphylococcus toxins (
1). Staphylococcal enterotoxins (
se) are single polypeptides of approximately 25 to 28 kDa, and most of them are neutral or basic proteins with isoelectric points (pIs) ranging from 7.0 to 8.6, with a high degree of microheterogeneity (
2). The stability of the
ses to proteolytic enzymes (trypsin, chymotrypsin, rennin, and papain), pH, heat, and gamma radiation was described by Bergdoll and Wong (
3). In a review published in 2007 by Bhatia and Zahoor (
4), the main elements of the pathogenicity of
S. aureus (toxins and invasiveness) have been discussed. Staphylococci esp.
S. aureus is highly correlated to nosocomial and community acquired infections. The pathogen is present as a part of the normal flora in humans, acting as a skin commensal. It may become pathogenic and provoke some skin infections and abscesses, or result in life-threatening diseases such as septicemia, mastitis, pneumonia, meningitis, endocarditis, urinary tract infections, toxic shock syndrome (TSS), phlebitis, endocarditis, and osteomyelitis (
4). Poisoning initiated by
se is one of the most common food-borne diseases worldwide resulting from the ingestion of
se pre-formed in food by enterotoxigenic strains of coagulase-positive staphylococci, mainly
S. aureus. The exact diagnosis of poisoning can only be based on the detection of
se in food (5). Twenty-one staphylococcal enterotoxins (
ses) have been identified: from
sea to
se1v. All have superantigenic activity whereas only a few (
sea to
sei,
ser,
ses and
set) have been detected to have emetic activity. These toxins can be produced only by enterotoxigenic strains of coagulase-positive staphylococci (mainly
S. aureus) in food with high protein content (
6).
The combined ability of
S. aureus to produce one or more enterotoxins and the methicillinresistance has been frequently reported in scientific publications of recent years. One of these studies report findings of
S. aureus strains with genes responsible for the production of enterotoxins and resistance to methicillin in beef, marine, and freshwater fish products (
7). In 2014, a study showed that about 75% of isolated
S. aureus strains demonstrated resistance against penicillin and ampicillin (
8). These data suggest that foodborne strains with increased antimicrobial resistance may significantly diminish the treatment efficacy of many other infectious diseases in animals and humans. This high prevalence of
se genes is evident in isolates from food matrices, and further studies of enterotoxin genes prevalence may contribute to a better understanding of the role of
S. aureus in food poisoning cases and outbreaks. At the same time, the authors have not reported strains that have the ability to produce enterotoxins and to be methicillin-resistant (
8). Data from 2013 showed that the retail samples of bovine meat can be contaminated by pathogenic bacteria during human contact, meat processing, and may present a source of methicillin-resistant
S. aureus (MRSA) for consumers and others who handle raw meat. This study does not report MRSA isolates with the ability to produce enterotoxins (
9).
In 2009,
S. aureus was detected in 19% (28/148) of tested isolates from different food products, which were both enterotoxigenic and oxacillin positive. Only one isolate had the
mecA gene obtained by PCR (
10). The study from 2011 demonstrated that staphylococcal isolates found in Louisiana pork and beef meats at retail possessed various enterotoxin gene and antimicrobial resistance profiles. In addition, vigilant food safety practices have to be implemented for staff who handles raw meat products to prevent foodborne infections and intoxications due to
S. aureus contamination (
11). Only 3 MRSA isolates from dairy products were detected in 227 colonies of
S. aureus, without enterotoxin presence (
12). Processing of food and handling is also a major problem leading to food poisoning and infection provoked by
S. aureus contamination. Strains isolated from all samples produced some virulence factors as hemolysin, coagulase, DNase, and enterotoxins. These strains were found to be resistant to several antibiotics. Ampicillin and penicillin were found to have the highest antimicrobial resistance (
13).
The aim of this article was to describe foodborne poisoning outbreaks (pig meatballs and potato salad) initiated by MRSA
mecA strains of
S. aureus with the ability to produce of wide range of staphylococcal enterotoxins.
MATERIAL AND METHODS
Epidemiological informationOn March 8, 2017 in the village Mamarchevo, the district of Yambol, information was received about a foodborne outbreak after the consumption of pig meatballs and potato salad. Two children (4-5-years-old) and eighteen adults (women, age betweeen 50-70 years) have shown signs of sickness – vomiting and diarrhea. All cases were immediately hospitalized. One female subject (76-year-old woman) was hospitalized with signs of hyperthermia (>38 °C), abdominal pain, vomiting, diarrhea, and fatigue.
S. aureus was isolated from the fecal sample. Epidemiological observations showed that the subjects mainly consumed potato salad and meatballs. These food products were prepared in uncontrolled conditions and were stored at room temperature for an extended period of time. Nasal and pharyngeal swab samples have been taken from three people included in preparing the food. Two samples were positive on
S. aureus with methicillin resistance. Food safety authority officials have collected samples from the prepared food. The food safety regional laboratory in Sliven detected
S. aureus in the meatballs and the potato salad at level 1x108 CFU/g. Samples of the food products and the
S. aureus isolates were sent to the National Reference Laboratory, NDRVMI in Sofia.
MethodsThe two samples (pig meatballs and potato salad) and the two
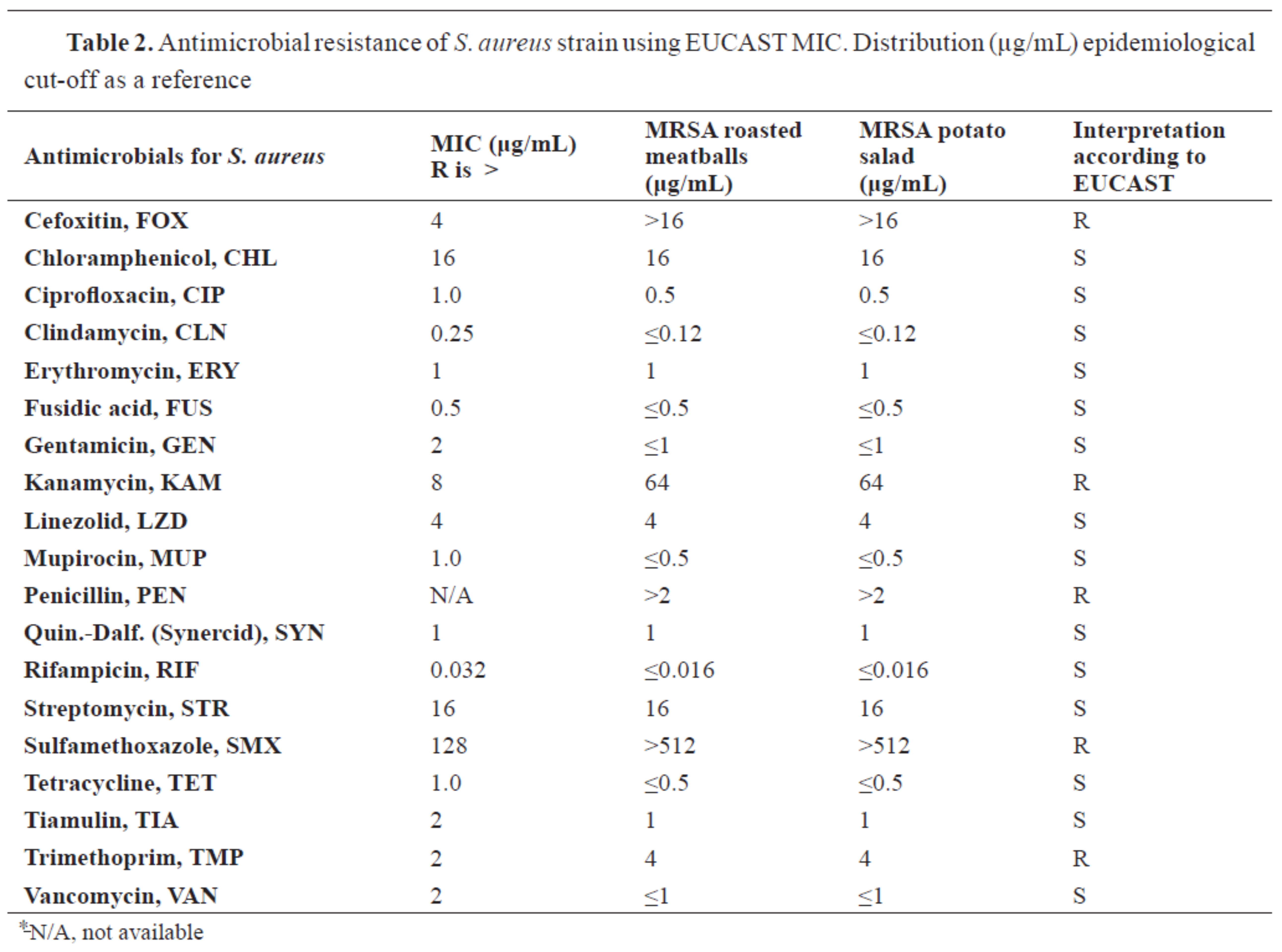
S. aureus strains from the same food matrices were subjected to a routine microbiological analysis for the presence of coagulase-positive staphylococci (CPS). Baird-Parker agar with the addition of egg yolk emulsion (Merck, Germany), incubated at 37 °C for 24–48 h was used to detect of CPS (ISO 6888 - 1/A1: 2005). All colonies showed the typical appearance of CPS, and they were tested for coagulase activity with rabbit plasma fibrinogen (Merck, Germany), catalase and antibiotic susceptibility analysis using the method of minimal inhibitory concentrations (MICs, mg/mL).
Isolation of CPS and identification of S. aureusIsolation of CPS was performed as follows: 10 g of each sample was diluted with 90 mL of Buffered Pepton Water (Merck, Germany) and homogenized in a stomacher (400 Circulator, England). Each sample was separated in 10-fold serial dilutions. A volume of 0.1 mL of the supernatant was spread in duplicate on Baird-Parker Agar plates (BPA) (Merck, Germany) and incubated under aerobic conditions at 37 °C for 24 and 48 h. The samples producing typical colonies (grey-black, surrounded by a dull halo) were considered to contain CPS (
14). The isolated strains were typical colonies of CPS. Additional tests for catalase and coagulase activity were performed. The coagulase test was performed with rabbit plasma fibrinogen (Merck, Germany) and Heart Infusion Broth (HiMedia, India).
Biochemical characterizationBiochemical identification of the isolates was performed using MICRONAUT-RPO plate (Merlin, Germany) for Gram (G) positive bacteria. The principle of the test is the addition of the bacteria suspension in saline to the microplate, which rehydrates the substrates dried onto the surface of the microplate wells. Following incubation at 37 °C for 24 hours the plate was scanned and analyzed using the MICRONAUT Scan under the control of the MICRONAUT software. Forty-four biochemical reactions were analyzed to calculate the identification profile.
Antibiotic susceptibility testingThe MIC for
S. aureus strains were determined by the broth microdilution method according to (
15). After 24 h at 37 °C, the inoculum was prepared of culture on CASO agar plates by suspending in sterile Ringer’s solution in order to obtain turbidity equivalent to 0.5 McFarland standards. The plate had the code EUST, Sensititre for G-positive microorganisms. Reference strain used for quality control was
S. aureus NBIMCC 3703 (LMG, Belgium-LMG 8224) = ATCC 25923.
Staphylococcal enterotoxins (ses) detection and quantificationExtraction and detection of ses using qualitative immunoassays were performed in NRL-Bulgaria. The CPS were analyzed in EURL (European Union Reference Laboratory), ANCES, France, according to the European screening method v5. Briefly, the samples received for analysis were first submitted to a protein extraction followed by dialysis concentration. The extracts were analyzed with a validated kit (Vidas SET2, bioMérieux®, bioMerieux, Marcy l’Étoile, France), which are able to detect
sea to
see simultaneously in dairy products (European Screening Method).
European Screening Method (ESM) positive extracts were submitted to the in-house quantitative enzyme-linked immunosorbent assay (ELISA) confirmatory method (ANSES) for
sea to
see characterization and quantification.
Quantification of
se was performed by a quantitative indirect sandwich-type ELISA. A single sandwich-type was used for
seb whereas double sandwich ELISA was used for
sea,
sec and
sed. Limit of Quantifications (LoQs) where estimated, as follow: 0.111 ng/mL for
sea, 0.128 ng/mL for
seb, 0.033 ng/mL for
sec and 0.182 ng/mL for
sed. This method was used in the frame of several studies on
ses detection in foods (
16,
17).
Staphylococcal enterotoxins genes (se) characterizationCPS were enumerated in suspected samples using the standard method EN ISO 6888 part 1 as described in the relevant EU legislation (Commission Regulation 2073/2005). CPS isolates were tested for enterotoxin genes by PCR targeting the
S. aureus 23S rRNA gene and biotyped as described previously (
18). The isolates were tested for
sea-e,
seg-j,
ser and
sep genes using two multiplex PCR assays according to the procedures of the EU Reference Laboratory (EU-RL) for CPS. The isolates were also typed by pulsed-field gel electrophoresis (PFGE) (
18).
Methicillin resistant Staphylococcus aureus genes characterizationProtocol for PCR amplification of
mecA,
mecC (
mecALGA251),
spa and
pvl, published by the EURL Antimicrobial Resistance, 2st Version, September 2012 (
19) was applied. The Protocol was developed using described method for multiplex PCR for detection of
mecA,
mecC (
mecALGA251),
lukF-PV (PVL) and
spa (
20).
RESULTS
The two samples (roasted pig meatballs and potato salad) and the isolates of CPS were received in the NRL of NDRVMI. Food samples had high levels of CPS. The value of
S. aureus contamination in potato salad was 8.3 logs CFU/g and 7.7 logs CFU/g in the roasted meatballs, which is significant to initiate production of toxin. Both
S. aureus strains were positive for catalase and coagulase, oxidase-negative immotile cocci with well apparent β-hemolysis on blood agar. MICRONAUT-RPO plate (Merlin, Germany) was used to confirm the presence of
S. aureus. The biochemical characteristics of the analyzed strains are presented in
Table 1.

The food samples were analyzed according to the European Screening Method v5 using mini VIDAS SET2 system. The results showed the presence of SE (TV 2.67 for the meatballs and TV 3.27 for the potato salad), which confirmed food poisoning. Results obtained in the NRL for the presence of
ses were re-tested in EURL CPS (European Union Reference Laboratory for Coagulase Positive Staphylococci), ANCES using the same matrices.
The MICs of
S. aureus strains were tested, using EUST sensititre plates. Resistance to cefoxitin and penicillin was detected proving phenotypic MRSA characteristics of the tested strains. All results for antimicrobial testing are presented in
Table 2.
EURL-CPS used quantitative ELISA method to prove the presence of
sea and
sed (0.124 and 0.221 ng/g, respectively) in the roasted pig meatballs and
sea,
sec, and
sed (0.379, 0.008 and 1.796 ng/g, respectively) in the potato salad.
In EURL CPS the two CPS isolates were further analyzed and characterized. These isolates were confirmed using the PCR test for
S. aureus by a species-specific 23S rRNA targeted PCR. Isolates from both matrices were analyzed for presence of se genes. The data showed that both strains were positive for the
sea,
sed,
seg,
sei,
sej, and
ser genes.
Also, the multiplex PCR method following the Protocol of EURL Antimicrobial Resistance was applied and the presence of
mecA gene was found (
Fig. 1).
Both isolates from roasted meatballs and potato salad were examined and they showed presence of the same gene.
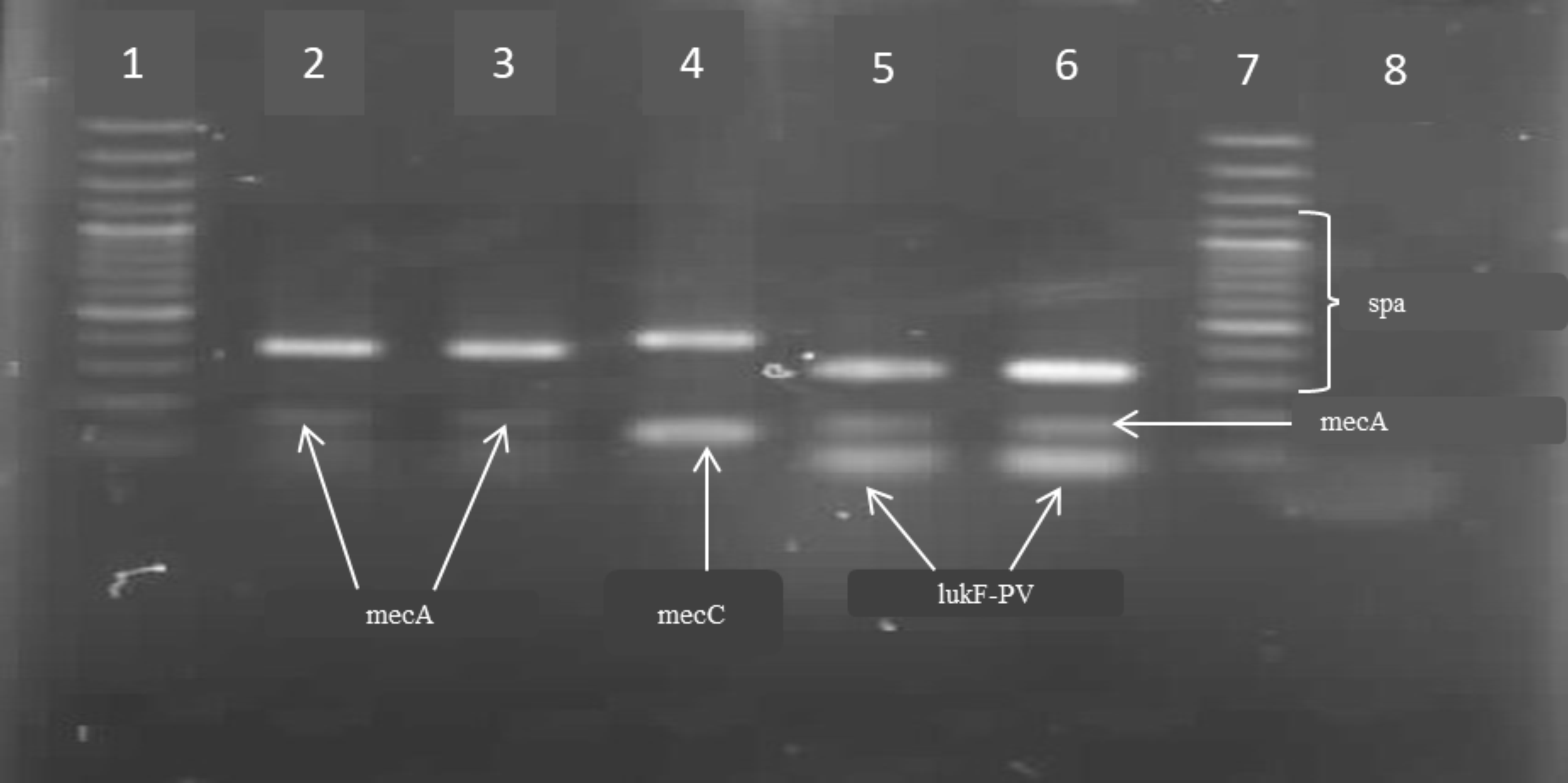 Figure 1.
Figure 1. Multiplex PCR method for detection of
mecA,
mecC (
mecALGA251),
lukF-PV (PVL) and
spa Legend: Lines 1 and 7: 100-bp ladder; Lane 2: MRSA roasted meatballs; Lane 3: MRSA potato salad; Lane 4:
mecC positive MRSA (
spa and
mecC amplification); Lines 5 and 6:
pvl positive MRSA (
lukF-PV,
spa and
mecA amplification); Line 8: negative control (H
2O)
DISCUSSION
Very few cases of foodborne poisoning outbreaks caused by
S. aureus strains are characterized by the ability of the pathogen to produce a wide range of enterotoxins and to demonstrate phenotypic methicillin resistance by the presence of the
mecA gene. The results of the current study describe food-borne (pig meatballs and potato salad) poisoning with the pathogenic strains of
S. aureus characterized by methicillin resistance (
mecA gene) and enterotoxin production encoded by six genes (
sea,
sed,
seg,
sei,
sej and
ser). Expression and secretion of toxins and enzymes of this array are tightly controlled by a number of regulatory systems.
S. aureus is also notorious for its capability to be resistant to a high number of currently available antibiotics and dissemination of various multidrug-resistant
S. aureus clones which limits therapeutic options for this type of infection (
21). The genetic association between antimicrobial resistance and enterotoxins is poorly understood. The evidence indicates that there is a significant correlation between the sea gene and both imipenem and ciprofloxacin resistance. Moreover,
sec gene was significantly associated with MRSA isolates. An interesting correlation was also found between high
sea gene expression and multidrug-resistance (
22). By comparing our results with this data, we can conclude that there is an unclear understanding of the relationship between genes corresponding to the enterotoxin production and genes related to methicillin resistance.
The occurrence of MRSA in retail foods in Shaanxi, China indicated that the pathogens could be from both animal and human origin despite the low prevalence (
22). The presence of multidrugresistant and enterotoxigenic MRSA strains in foods poses a potential threat to consumers and emphasize the need for better control of potential sources of contamination (
23). Our findings proved the possibility that
S. aureus strains are able to combine methicillin resistance and multiple enterotoxin production encoded by several genes. Some new approaches as phylogenomics of
S. aureus, using whole-genome surveillance will replace other forms of molecular typing, providing data for understanding the evolutionary dynamics (
24). Pulsed-field gel electrophoresis (PFGE) can yield data for various strains with antimicrobial resistance, producing enterotoxins and hemolysins.
In a study by Alibayov et al. (
8), 82.8% (n=93) of the isolated
S. aureus strains, harbored one or more of the following enterotoxin genes (
sea,
seb,
sec,
sed,
see,
seg,
seh,
sei,
sej); 39.8 % of the strains demonstrated
se genes and 43% carried from two to five
se genes of the genes examined. The most commonly detected toxin genes were
sea,
seb,
sec and
seg. The presence of genes coding for antibiotic resistance such as
mecA was investigated by PCR. Ten strains (10.75%) carried the
mecA gene and correspondingly demonstrated resistance to oxacillin (
8).
In our study, both
S. aureus strains contained
mecA gene and
sea,
sed,
seg,
sei,
sej and
ser genes. Our results demonstrate that
S. aureus can have both enterotoxin-producing and methicillinresistance genes.
A study by Pu et al. (
25), characterized 152
S. aureus strains, including 22 MRSA collected from Louisiana retail pork, for the prevalence of nine enterotoxins and four other exotoxin genes by PCR method and antimicrobial susceptibility testing by broth microdilution. Overall, 85% of
S. aureus isolates were positive for at least one of six enterotoxin genes identified. The most predominant genes were
seg and
sei (66% each), followed by
seh (20%),
sed (15%),
sej (13%), and
sea (1%). Resistance to penicillin (71%), ampicillin (68%), and tetracycline (67%) was common, followed by erythromycin (30%), clindamycin (18%), oxacillin with 2% NaCl (14%), ciprofloxacin (13%). Multidrug resistance was commonly observed among MRSA isolates and
S. aureus isolates from pork (
25).
In our study, we detected six enterotoxigenic genes and antimicrobial resistance to cefoxitin, kanamycin, penicillin, trimethoprim and sulfamethoxazole. Researchers reported one MRSA strain with unusual characteristics: it was oxacillin susceptible, harbored SCC
mecV, and was positive for
sed,
seg and
sej (
26). In a study by Jackson et al. (
27), staphylococci isolated from retail pork and beef in Georgia, were compared to MRSA strains collected from clinical patients from the same geographic area, using broth microdilution antimicrobial susceptibility testing, multilocus sequence typing (MLST),
spa typing, SCC
mec typing, and PFGE. Multidrug resistance was detected among MRSA from all sources. This data suggests that the retail meat samples were contaminated by a human source, possibly during the processing of the meat, and may present a source of MRSA for consumers and others who handle raw meat (
27). Our study proved the same relationship between personnel who cooked the food (potato salad and meatballs) and ill consumers.
As early as 1991 а combination of enterotoxin production and methicillin resistance of some
S. aureus isolates was noted (
28). In our study, we have detected six genes for the production of enterotoxins in combination with the presence of
mecA. In a study by Kamarahei et al. (
29), a high proportion of
S. aureus isolates from Iran carried
sea gene (60.6%), whereas the frequency of
seb gene in North Iran was less distributed than all strains (27.1%). Some strains had the following combination of genes –
sea,
seb and
mecA or
mecC (
29). Food producers may play the role of a reservoir of virulent strains of
S. aureus and may be vectors of transmission to food. In a study by Santos et al. (
30), the prevalence of
S. aureus was 19.8% in the nose and 11.1% on the hands of the workers; 6.2% of the individuals carried
S. aureus both in their noses and hands, and three persons had the same strain (PFGE type) in the nose and on the hands. Although 82% of the isolates were resistant to at least one antibiotic, none demonstrated the presence of either
mecA gene or resistance to oxacillin (none identified as MRSA). Sixty-eight percent of the isolates from the nose and hands possessed enterotoxin genes (
30).
MRSA as a food-borne pathogen is a publichealth threat considering the prevalence in domestic animals, foods of animal origin and consumers, and their ability to produce enterotoxins. Research data indicate that
S. aureus with enterotoxigenic characteristics plus methicillin resistance can also act as food-borne pathogens upon favorable conditions for growth and enterotoxin production. The degree of the intoxication is not related to the antimicrobial resistance profile of the causative
S. aureus strain and therefore MRSA food-borne outbreaks are not expected to be more severe (
31).
CONCLUSION
In conclusion of the discussion of the obtained results and their comparison with similar studies of the specificity of
S. aureus isolates found in our research, we can summarize the growing importance of such combination of different pathogenic elements (six genes for enterotoxin production plus
mecA gene for MRSA resistance). The presence of genes that determine the production of several
se in combination with pronounced methicillin resistance makes such strains extremely dangerous to humans, regardless of the mode of their transmission. The presence of genetic structures that cause multiplied pathogenicity poses new challenges in the fight against
S. aureus, regardless of the mode of transmission of the living causative agent and the products of its vital activity (enterotoxins).
CONFLICT OF INTEREST STATEMENT
All authors declared that they do not find any potential conflict of interest with respect to the authorship and/or publication of this paper.
ACKNOWLEDGEMENTS
All authors express their thanks for help of Yacine Nia and Frédéric Auvray from EURL CPS for examining the type of staphylococcal enterotoxin trough quantitative ELISA method and for the detection of staphylococcal enterotoxin (se) genes.
AUTHORS‘ CONTRIBUTIONS
TI participated in isolation, identification, AMR testing, PCR study, writing the article. GK-V participated in PCR study and literature review. GM performed miniVIDAS testing and PCR study. HD carried out supervision of all processes of testing, writing the paper and adaptation to the journal.

 10.2478/macvetrev-2020-0026
10.2478/macvetrev-2020-0026


