Bovine rotavirus A (BRVA) is a frequent causative agent of diarrhea in neonatal calves. Accurate and rapid diagnosis is crucial to prevent calf mortality from BRVA induced diarrhea. Currently, variety of diagnostic methods are being used to detect BRVA from calves’ feces: antibody-based rapid test and ELISA, and molecular-based RT-PCR and RT-qPCR. The aim of the study was to evaluate the accuracy (sensitivity and specificity) of the rapid test (Immunochromatography), ELISA, and RT-PCR assays, using RT-qPCR as the gold standard, in detection of BRVA in diarrheic calves’ fecal samples. One hundred (n=100) clinically diarrheic fecal samples were tested with four different diagnostic tools. The percent of samples positive by rapid test, ELISA, RT-PCR and RT-qPCR was 10%, 16%, 17%, and 33%, respectively. The agreement between different assays was 75% to 99%. The highest agreement was observed between ELISA and RT-PCR assay (99%). The lowest agreement was recorded (75%) between rapid test and RT-qPCR. The sensitivity of the rapid test, ELISA, and RT-PCR were 30%, 49%, and 52%, respectively when compared to the reference test (RT-qPCR), whereas specificity was 100% for all assays. In conclusion, none of the frequently used diagnostic tests showed a satisfactory level of sensitivity to identify BRVA in calves’ feces. Therefore, the use of a more sensitive rapid test should be used to identify infected calves in field conditions in order to prevent calf mortality from rotaviral diarrhea.
Diarrhea is one of the most common and threatening health problems in dairy calves worldwide (
1). It plays a vital role in morbidity and mortality of dairy calves younger than 6 weeks of age and subsequently causes potential losses in the dairy industry (
2). Bovine rotavirus (BRV), Coronavirus (BCV),
Escherichia coli (K99),
Salmonella strains, and
Cryptosporidium parvum are more frequently reported pathogens responsible for neonatal calf diarrhea (
3,
4). Rotavirus itself is accounted for 27-36% of calf diarrhea (
5). It is clinically associated with liquid diarrhea in 9 to 21-day old dairy (
3) and beef calves (
2). BRVA is the most common pathogen in the environment, and comparatively to other pathogens, highly resistant to numerous disinfectants (
6). It spreads mostly via a fecal–oral route through animal-toanimal contact or consumption of contaminated feed (
7). Usually, adult animals are the prime source of infection for newborn calves (
6). It is well established that diarrhea is a complex and multi-factorial clinical symptom, so specific diagnosis is very difficult without confirmatory tests. Moreover, early detection of the causal agent is crucial so that rapid treatment and prevention strategies can be executed. Diagnosis of BRVA infection depends on the detection of viral particles in stool samples obtained early in the course of the clinical manifestation (
8).
Rotaviruses belong to the family of
Reoviridae, genus
Rotavirus. The icosahedral non-enveloped viruses possessing genome is composed of 11 double-stranded RNA (dsRNA) segments that encode six viral structural proteins (VP1 to VP4, VP6, and VP7) and six nonstructural proteins (NSP1 to NSP6) (
8). Among them, NSP4 acts as a viral enterotoxin exerting both secretory and subsequent anti-secretory actions which result in moderate loss of Cl- into the intestinal lumen at the onset of BRV diarrhea (
9). The virus capsid contains three concentric protein layers, including an outer layer composed of the VP7 and VP4 proteins, a middle VP6 glycoprotein layer, and the central core-shell formed by VP2 (
6). The genus
Rotavirus has been divided into 8 groups or serogroups (A-H) based on antigenic epitope analysis of the VP6 glycoprotein (
10). Traditionally, viral classification has been performed based on serological characteristics and sequence diversity of the outer capsid proteins, VP7 (glycosylated, G-type) and VP4 (protease-sensitive, P-type) (
11). At least 35 G types and 50 P types of BRVA species have been detected up to this date (
12).
There are many tests applied routinely in diagnostic laboratories for the detection of BRVA in fecal samples. Electronic microscopy, polyacrylamide gel electrophoresis (PAGE), reverse passive hemagglutination, immunochromatography (IC), Enzyme-linked immunosorbеnt assay (ELISA), and more recently, molecular techniques such as RT-PCR and RT-qPCR have replaced other diagnostic tests with the advantage of higher analytical sensitivity and specificity (
6).
When outbreaks of diarrhea occur in a herd, immediate identification of virus-associated diarrhea is obligatory to ensure the administration of appropriate treatment and control. For this reason, the selection of rapid and sensitive diagnostic tools is mandatory. The immunochromatography (IC) strip test is a rapid calf-side pathogen detection test for assessment of fecal samples in the field; on the other hand, ELISA, RT-PCR, and RT-qPCR require laboratory conditions. The sensitivity and specificity may be variable between diagnostic tests. Evaluation of diagnostic tests in terms of sensitivity, specificity, and estimation of predictive values could be very useful in selecting a more reliable diagnostic tool. The use of inaccurate diagnostic tests might cause underestimation of disease burden in a population and would lead to increased mortality from a nonspecific and delayed therapeutic response. A limited number of studies have been conducted to evaluate different diagnostic tests for rotaviral infection in calves; none recorded in Bangladesh. Therefore, in the present study, we aimed to compare and confirm a rapid and/or reliable diagnostic test for the diagnosis of rotaviral cases in calves using fecal samples.
MATERIAL AND METHODS
Sample collectionSamples were collected from different dairy farms in the Chattogram division of Bangladesh in the period from July 2015 to May 2016. A total of 100 fecal samples were aseptically collected from the rectum by gloved-finger method from acutely diarrheic calves aging between 1 and 45 days. Precautionary measures were taken so as to avoid contamination during sample collection, transport, storage, and processing. Immediately after the collection, the samples were aliquoted, stored in a cool box containing an ice pack (4 °C), and transferred to the laboratory.
Sample processingOn the day of collection, the samples were transported to the clinical pathology laboratory in the Department of Pathology and Parasitology, Chattogram Veterinary and Animal Sciences University. We divided each fecal sample into 2 ml aliquots, the first being stored at 4 °C for ELISA testing, and the second 0.1 g of undiluted feces was mixed with 0.9 ml phosphate-buffered saline (PBS), and then stored at 4 °C for the RT-PCR assay. The remaining fecal samples were stored at -70 °C for further testing, if required. All the samples (N=100) were repeatedly tested using four (
4) diagnostic tests for BRVA detection.
Lateral flow immunochromatography (Rapid test)Within a short time of samples reception at the laboratory, LAT dipsticks (Bio-X® Diagnostics; Jemelle, Belgium) were used for the detection of BRVA following the manufacturer’s instructions. Briefly, a small amount of feces was homogenized in a buffer solution and the dipstick was placed into the suspension. A sample was considered positive when both the control and positive indicator lines turned red and negative if only the control indicator line turned red. If the control indicator line failed to turn red, the test was regarded null (indicative of a faulty dipstick) and was retested using another dipstick.
Enzyme-linked immunosorbent assay (ELISA) testWe used a commercial ELISA kit for rotavirus (Bio-X® Diagnostics; Jemelle, Belgium) and performed the test in accordance with the procedure described by Barua et al. (
5). Dilution buffer (50 μl) was mixed with undiluted feces samples (50 μl). The mixture was placed into the wells of a microplate coated with the corresponding antibody. The plate was kept at room temperature (approximately 21 °C) for 60 minutes followed by a manual wash using the washing-solution. The conjugate solution (100 μl) was added to the wells for each sample and the plate was held at room temperature (approximately 21 °C) for another 60 minutes. After the final wash, tetramethylbenzidine (TMB) substrate was added to each well and the plate was incubated at room temperature for a further 10 minutes. Then, a stop solution (50 μl) was added, and the optical densities (OD) were measured at 450 nm using an ELISA plate reader (Labsystems Multiscan Biochromatic; Labsystems, Basingstoke, UK) (
13). The OD was measured, the values were transformed into Microsoft excel for calculation of the S/P ratio, and further calculations were performed according to the manufacturer’s recommendations.
RNA extraction for RT-PCR and RT-qPCRFor RNA extraction we used a magnetic beadbased system (MagMax 96 Viral RNA, AM 1836 Ambion, Austin, TX, USA) in accordance with the manufacturer’s instructions and methods described by Barua et al. (
5). Initially, a 10% suspension of fecal samples was prepared in PBS and centrifuged with low speed (1500 g, 4 °C for 10 minutes). Using a magnetic particle handling system (Kingfisher 96, Thermo, Finland), the magnetic beads were handled and washed, and the nucleic acid was eluted. The 50 μl eluted nucleic acid was stored frozen at -20 °C until tested. The RNA was denatured by heating at 95 °C for 5 minutes before testing for BRVA using RT-PCR.
RT- PCRThe primers used for amplifying a 294-bp fragment of the VP6 gene of BRVA, the sequence of the upstream primer was 5’-ACCACCAAATATGACACCAGC-3’; the sequence of the downstream primer was 5’-CATGCTTCTAATGGAAGC-3’ (
14). For RT-PCR we used Superscript One-Step RT-PCR System (Life Technologies, Rockville, Md.) following the manufacturer guidelines. 5 μl of extracted RNA 0.4 μl M primers were mixed together, and RNase-free water was added up to a total volume of 24 μl. At 95 °C mixture was heated for 5 min and then quickly cooled to 4 °C. Superscript 2 X-reaction mixes (25 μl) and RT-
Taq mix (1 μl) were then added. The Cycling conditions of PCR were as follows: (a) reverse transcription for 30 minutes at 50 °C; (b) a 15-min activation step at 95 °C; (c) 40 cycles of 30 seconds at 94 °C, 60 seconds at 55 °C, and 60 seconds at 72 °C; and (d) final extension for 7 minutes at 72 °C (
15). For each assay we used one negative and one positive control (derived from known rotavirus positive samples). The amplified products were analyzed by electrophoresis on 1% agarose with ethidium bromide-stained gels, and then photographed by a gel documentation system (
Fig. 1).
 Figure 1.
Figure 1. RT-PCR amplification of VP6 gene. The PCR amplification of VP6 gene of BRVA, showing positive amplicons at 294 bp M: DNA size marker (100–1000 bp), Lane P: Positive control, Lane N: Negative control, Lane (1-7): VP6 gene positive isolates
Real time RT- PCRThe RT-qPCR assay was performed by using an ABI 7500 thermocycler (Applied Biosystems). The following TaqMan® assay primers and probe sequences and thermal cycles were selected for testing according to the previously described method by Jothikumar et al. (
16): forward primer JVKF (5¢-GTGGTTGATGCTCAAGATGGA-3¢; positions 17–39), reverse primer JVKR (5¢- TCATTGTAATCATATTGAATACCCA-3¢; positions 147–123), and TaqMan® probe JVKP (FAM-5¢-CTGCAGCTTCAAAAGAAGWGT-3¢ Black Hole Quencher; positions 96–72). Prior to the addition of RNA to the RT-PCR master mix for separation of the rotaviral dsRNA, samples’ RNA was denatured by heating at 95 °C for 5 minutes followed by incubation in ice for 5 minutes. The one-step rotavirus assay was performed in a 96-well plate format. One-step RT-qPCR amplifications were performed using the following conditions: reverse transcriptase reaction for 30 minutes at 50 °C, followed by denaturation at 95 °C for 15 minutes, followed by 45 cycles of denaturation at 94 °C for 10 seconds, annealing at 55 °C for 30 seconds, and extension at 72 °C for 20 seconds. Two negative and two positive controls were used (derived from known rotavirus positive samples) (
Fig. 2).
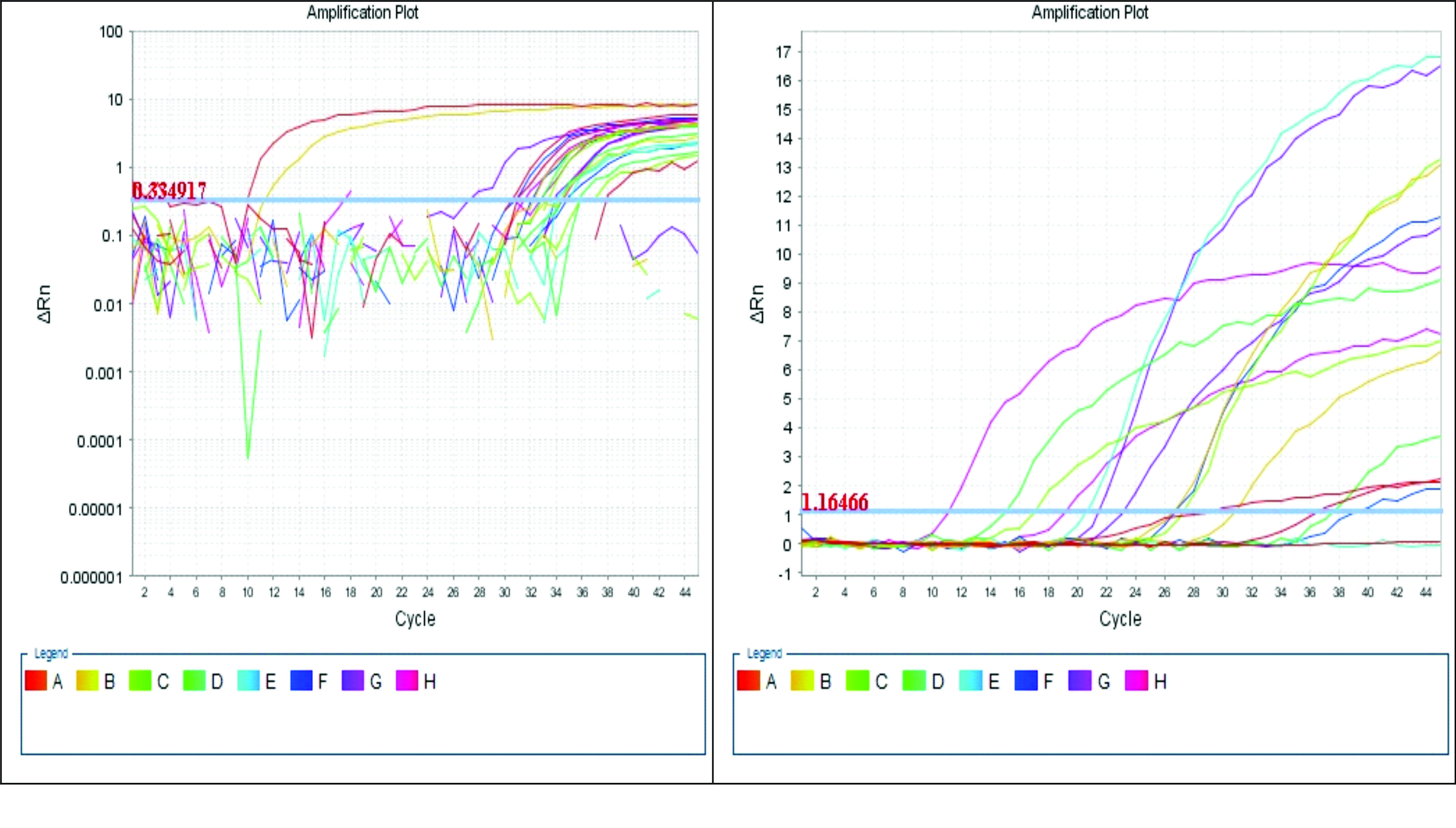 Figure 2.
Figure 2. Amplification of cyclic reaction in RT-qPCR
Test evaluationSensitivity, specificity, positive and negative predictive values of the different tests were determined by its’ comparison with the RT-qPCR as a reference test. RT-qPCR and ELISA were compared according to viral load based on the ELISA S/P ratio and RT-qPCR Ct values. A higher ELISA S/P ratio and a lower RT-qPCR Ct value were indicative of higher viral concentrations. The ELISA S/P ratio results were divided into four categories: <7 (negative), 7–25, 26–70, and >70. The Ct values for the RT-qPCR were divided into four categories: 5-15, 16-25, 26-35, 36-40. The agreement between different tests was tested by kappa statistic. The kappa (k) value was interpreted as one of the following: poor (k=0), slight (0.01<k<0.20), fair (0.21<k<0.40), moderate (0.41<k<0.60), almost perfect (0.61<k<0.80), and excellent (0.81<k<1.00) (
17).
Statistical analysisAll the collected data were entered into MS excel (Microsoft office excel-2007, USA). SPSS 20.0 Statistics software was used to calculate the percentage of the agreement between the different diagnostic assays and Cohen’s Kappa coefficient and the agreement was determined following Carrouel et al. (
17). For sensitivity, specificity, positive predictive value and negative predictive value were calculated mathematical formula described by Shaha et al. (
18).
RESULTS
One hundred fecal samples of diarrheic calves from selected areas of Chattogram, Bangladesh, were submitted to clinical pathology laboratory and subsequently tested by rapid test, ELISA, conventional RT-PCR, and RT-qPCR for detection of BRV antigen. From the total number of analyzed samples (N=100), 10%, 16%, 17%, and 33% were positive according to the rapid test, ELISA, RT-PCR, and RT-qPCR, respectively (
Table 1). Ten samples were positive for rotavirus in all diagnostic assays.

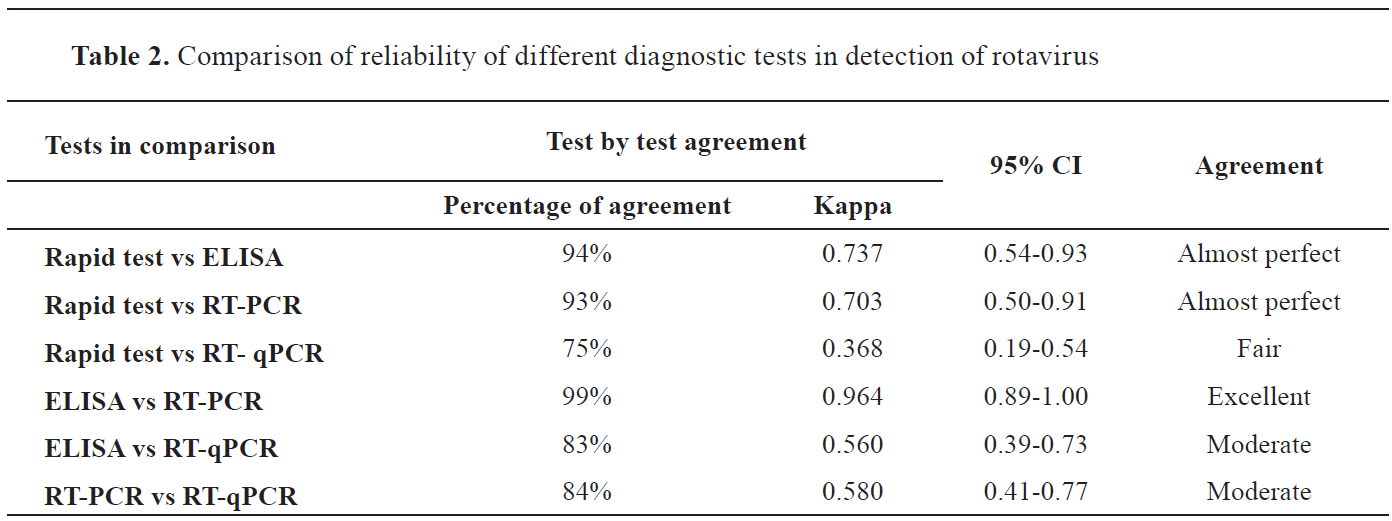
The rapid test showed almost perfect agreement with ELISA and RT-PCR results. Fair agreement was observed between rapid test and RT-qPCR. Similarly, ELISA and RT-PCR had excellent agreement, but ELISA showed moderate agreement with RT-qPCR. Moreover, we also compared the RT-PCR with RT-qPCR and found moderate agreement among them. All the results of agreement between the different tests are shown in
Table 2.
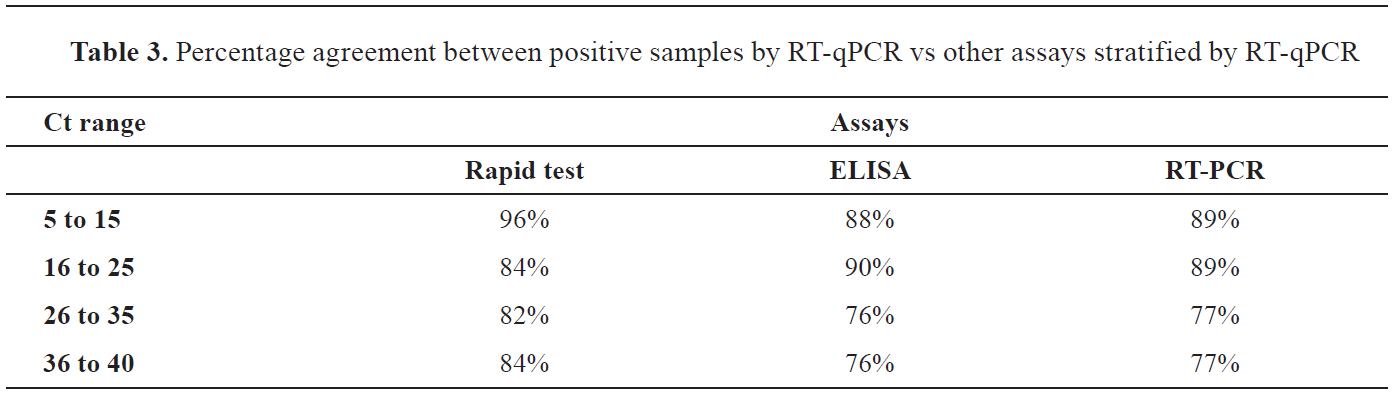
The Ct values of RT-qPCR test was inversely correlated with the values in the other three assays (
Table 3). The lowest Ct range 5-15 of RT-qPCR was correlated with the highest percentage of positive findings according to the rapid test, ELISA and RT-PCR. Similarly, highest Ct range 36-45 was correlated with the lowest percentage of positive findings according to the rapid test, ELISA, and RT-PCR.
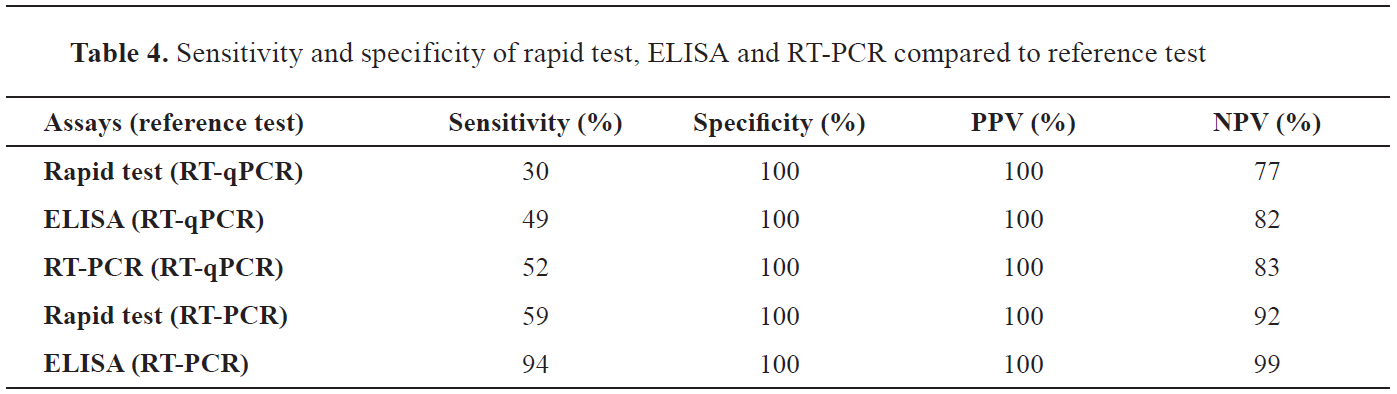
The comparison of the utilized tests with the reference “gold standard” test (RT-qPCR) showed fluctuation in sensitivity, though specificity was optimal (100%) (
Table 4). The sensitivity of the rapid vs the RT-qPCR and the RT-PCR was 30% (lowest among all) and 59%, respectively. Similarly, ELISA vs RT-qPCR and RT-PCR showed 49% and 94% sensitivity, respectively. The RT-PCR vs RT-qPCR sensitivity was 52%.
Moreover, BRVA was detected by RT-qPCR in 18% of the samples that tested negative by using ELISA (S/P ratio <7). Furthermore, when the S/P ratio was >70, 58% samples had RT-qPCR Ct range within 5-15 (
Fig. 3).
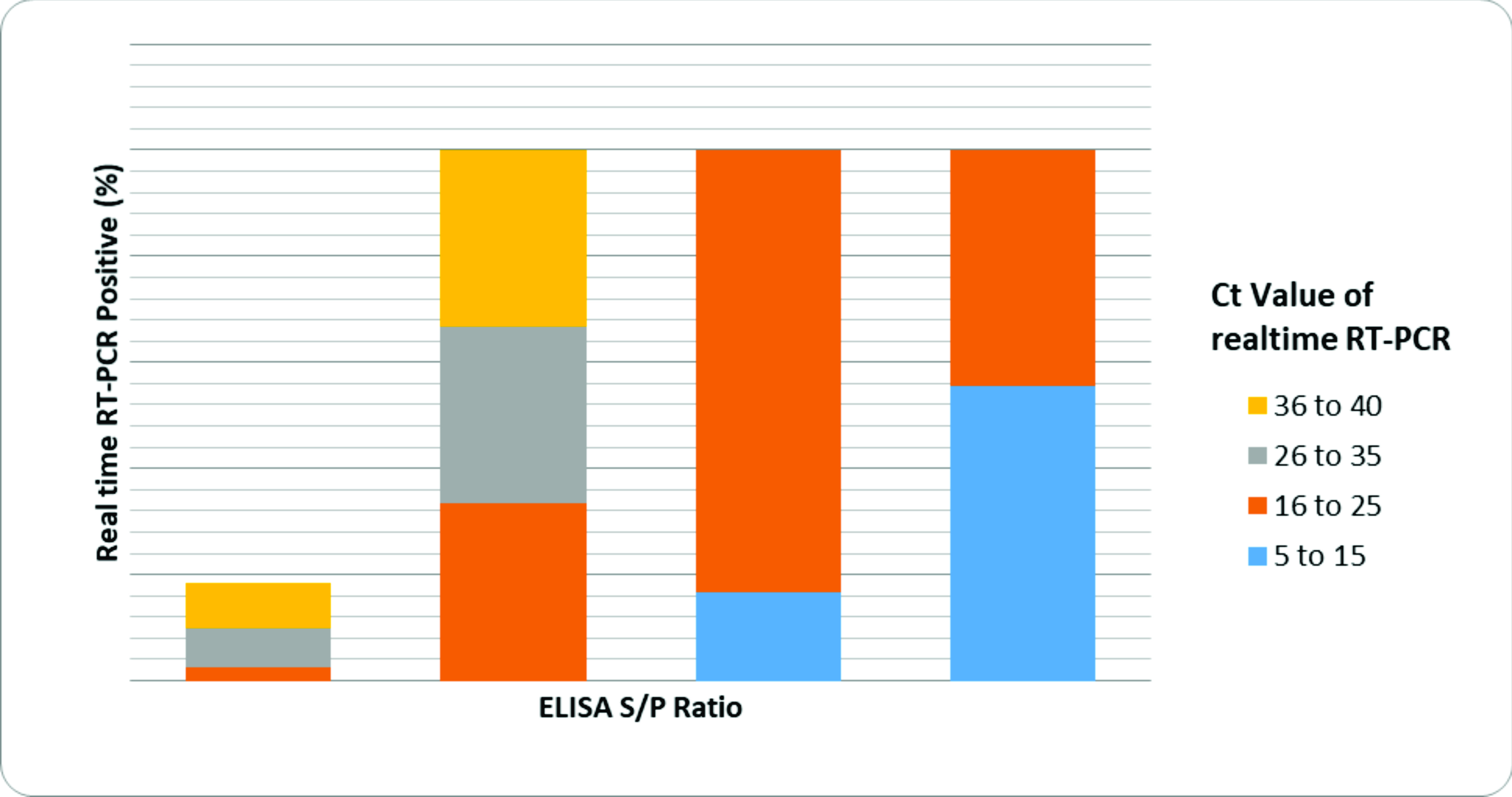 Figure 3.
Figure 3. Relationship between ELISA S/P ratio and real time RT- PCR cycle threshold (Ct) for samples tested for BRVA. The percentage of samples positive by RT-qPCR is demonstrated for each S/P ratio. ELISA S/P ratio <7 indicates negative test result. RT-qPCR Ct value: the lower the range, the higher the viral concentration
DISCUSSION
A simple, rapid, sensitive, and specific diagnostic technique is mandatory for the detection of viral agents causing gastroenteritis to provide appropriate on time treatment. Due to the multifactorial nature of the disease, it is essential to accurately identify the pathogen (quantitative assays) and to estimate its load and shedding from the affected animals. However, for small scale farm owners of developing countries, the use of quantitative techniques for diagnostic purposes, such as the RT-qPCR, would not be economically feasible. These farmers need a quick, less expensive and reliable diagnostic test. Regarding this, four routinely used diagnostic assays for the detection of rotaviruses in fecal samples from clinically diarrheic calves in Bangladesh were evaluated.
As rotavirus is one of the most important agents associated with severe diarrhea in humans and animal species, various methods have been developed for its antigen detection in fecal samples (
19). The sensitivity and specificity of different diagnostic tools are variable affecting the estimation of disease burden in a population. There are many factors affecting the choice of protocols used for diagnostic testing, including cross reactivity of different strain of targeted virus or bacterial pathogens, laboratory capacity to test a sufficient number of specimens per day, and type of the rotavirus circulating in the target population (
20).
The methods of choice for the detection of rotavirus in fecal samples should have high degrees of sensitivity and specificity, high predictive values and reproducibility (
21). In this study, we evaluated three potential frequently used diagnostic assays, rapid test, ELISA, and RT-PCR which were compared with the RT-qPCR for the detection of BRVA from diarrheic calf feces. We estimated the rate of positive samples for rapid test, ELISA, RT-PCR, and RT-qPCR as 10%, 16%, 17% and 33%, respectively. The detection rate of rotavirus from feces by antibody-based tests such as rapid test was found lower than the molecular techniques like RT-PCR and RT-qPCR in the study. However, the detection rate of antibody-based test ELISA and molecular test RT-PCR was almost similar. Usually, the main limitation of antibody-based tests is cross-reaction between different types of BRVA and coronaviruses; and the requirement of high concentration of antigen for getting positive results (
6). On the other hand, molecular based antigen detection techniques were found more effective for diagnosing the rotavirus from the clinical samples in previous studies (
22). However, in our study we observed almost similar result for ELISA and RT-PCR (agreement 99%). Even though different studies indicate variation of effectiveness of different diagnostic tools for the detection of rotavirus, our study clearly proved that molecular techniques, especially RT-qPCR is highly effective in comparison to other tests we applied. The RT-qPCR detected BRVA in some samples which were not detectable by other diagnostic methods applied. Similar findings also reported from other research and they found RT-qPCR as powerful diagnostic tool for routine detection of rotavirus in stool samples (
23). Besides, RT-qPCR can minimize or prevent the cross-reaction with other similar pathogen or particles.
Rapid test would be a very effective cow side diagnostic test because of its simple application and quick result. Rapid test has a number of advantages, including its simple format, rapidity, low-cost, and it can be performed without trained personnel or specialized laboratory equipment (
24). Moreover, it can be interpreted without using equipment, thus making it easily usable for any laboratory or farm. In our study we estimated remarkably low sensitivity (30%) of the rapid test in comparison to other tests. However, there is evidence that test results may vary depending on the rapid-test brand and that in some instances, they may be more accurate than the ELISA (
14). In the current study, the rapid assay was compared to the gold-standard tests, RT-PCR and RT-qPCR due to their high sensitivity and specificity (
25). The rapid assay sensitivity was 30% and 59% compared to RT-qPCR and RT-PCR, respectively.
In our study, 10 samples were positive for rotavirus by rapid test. However, when tested by RT-qPCR, rotavirus infection was detected in 33 samples. The relative prevalence of the RT-qPCR was almost four times higher compared to the rapid test. The low agreement (k=0.368) between the rapid test and the RT-PCR might be due to the lower sensitivity (30%) of the rapid test. However, specificity was 100% for both. Since, nucleic acid-based assays are generally much more sensitive than antibody-based assays for antigens (
24), the observed result of RT-qPCR for the detection of BRVA in feces may be more acceptable as diagnostic tool. But it should be noted that RT-qPCR needs laboratory equipment, skilled personnel as well as highly costly materials; whereas rapid test can be implemented on the spot. Therefore, a more sensitive rapid test would be the best choice for a quick, less expensive and appropriate detection. ELISAs are now commonly used in many diagnostic laboratories for the detection of rotavirus. Most of the ELISA tests have been developed for human rotavirus detection; some ELISAs have been adopted for veterinary medicine (
26). Very recently, ELISA assay kits have been developed for the detection of rotavirus from animal samples. ELISA test has a noteworthy limitation in generating positive results; it needs viral load 104-106 particles/ml of feces (
27). The detection limits of the ELISA test are at least 100- fold lower than that of the RT-PCR (
28). ELISAs are widely used in diagnostic laboratories because they provide rapid detection of rotavirus antigen in a relatively short time in comparison to other tests, like RT-PCR, RT-qPCR, and virus isolation (
6). ELISA is also good for handling large numbers of specimen at a time; however, it was found much less sensitive than RT-qPCR in our study. ELISA’s sensitivity was estimated 49% compared to the RT-qPCR in this study.
The true positive of a diagnostic test can be affected by other factors besides its sensitivity, such as the time of sample collection. Therefore, the optimal time for sample collection would be the clinical phase of the disease. It has been determined that during the early stages of the rotavirus-induced diarrhea, the viral load can be 109 particles per gram of feces (
11). Magnitude of virus shedding based on the Ct values of RT-qPCR can help to determine the cause of the disease (
6). High viral load in the samples help the clinicians to diagnose the etiology of the disease. A human based study revealed the correlation between the Ct values and the clinical stage of the disease. Ct values £25 was found associated with the diarrhea in the clinical stages of the disease. Rotavirus-A is shed typically in very high levels in the feces of rotavirus-A gastroenteritis cases (
29).
CONCLUSION
In order to identify and confirm a rapid and accurate diagnostic method for on-farm detection of pathogen causing diarrhea, four diagnostic assays were evaluated by comparing its performances in detecting BRVA in calf fecal samples. According to the measured sensitivity of different frequently used diagnostic tests, ELISA showed the highest sensitivity and specificity to detect infected calves in the herd. Rapid test showed highest percentage of agreement with RT-qPCR at different ct values. Hence, the use of rapid test would be the best choice for on-the-spot application. A highly sensitive and easily applicable diagnostic kits should be developed in the future.
CONFLICT OF INTEREST
The authors declared that they have no potential conflict of interest with respect to the authorship and/or publication of this article.
ACKNOWLEDGEMENTS
We thank the University Grants Commission of Bangladesh for their support through the Higher Education Quality Enhancement Project (CP: 3220). We thank Tofazzal Md. Rakib for his technical assistance during laboratory testing of the samples.
AUTHORS' CONTRIBUTION
SRB was responsible for conceptualization, data curation, formal analysis, investigation, methodology, writing original draft, writing review and editing. SI did the formal analysis, wrote the original draft, wrote the review and edited the paper. AZS, MM and MAH were included in the writing of the review and editing. SC was responsible for the conceptualization, formal analysis, funding acquisition, project administration, supervision, writing the review and editing.

 10.2478/macvetrev-2020-0033
10.2478/macvetrev-2020-0033






