The primary objective of our study was to detect the occurrence of enterotoxigenic Staphylococcus aureus in diverse types of cheese (cow’s milk cheese and mixed milk cheese) samples from R.N. Macedonia. Cheese samples were analyzed for enumeration and isolation of the S. aureus strains according to ISO 6888-1. We detected the toxigenic potential of the strains by the use of the Enzyme Link Fluorescent Assay VIDAS system, and we confirmed the presence of the SEs (sea, seb, sec, sed, see) genes by multiplex PCR. The results showed that out of 270 samples of cheese, coagulase-positive Staphylococcus (CPS) were detected in 27 (10%), and coagulase-negative staphylococci in five samples (1.8%). Biochemically, all 27 CPS samples were confirmed to be Staphylococcus aureus. With VIDAS SET2 test we confirmed that 11 isolates are producers of one of the toxins limited by the test. With the conventional PCR we confirmed genes in only 7 isolates. Most common detected gene was seb n=3 (42.8%), followed by sea n=2 (28.6%), and sec n=2 (28.6%). Additionally, sed and see genes were not detected in any of the S. aureus isolates. Discrepancies between the two test methods for detection of enterotoxigenic potential are not uncommon. The presence of viable Staphylococcus aureus cells that have enterotoxin potency demonstrates the importance of appropriate hygiene practices in the diary process and also the maintenance of the products in order to obtain a safe final product for the consumers.
Staphylococcal food poisoning (SFP) is one of the most frequent foodborne illnesses resulting from the ingestion of contaminated food with staphylococcal enterotoxins (SE) mainly produced by enterotoxigenic strains of
Staphylococcus aureus (
S. aureus).
Staphylococcus is accounted for the numerous food outbreaks worldwide. It produces various pathogenic factors such as enzymes, cytotoxins, exotoxins, and exfoliative toxins. The production of various toxins and the ability to produce biofilm are the two main virulent factors of
Staphylococcus spp. that affect its pathogenesis. Currently, 25 SEs are described (SEA – SElZ), not including the variants and TSST-1 (formerly known as SEF), and new types are frequently being discovered (
1). The SEs are powerful non-specific T-cell stimulators (superantigens) that cause unregulated activation of the immune response. The enterotoxins which demonstrated emetic potential in monkeys were designated as “SE”, whereas the others that were not evaluated or were confirmed to have no emetic effect are designated as enterotoxin-like toxins (SE
l-), with exception of TSST-1.
Food can be easily contaminated by
S. aureus as well as with other enterotoxigenic strains which is a potential risk to public health. Raw milk and cheese are considered as the most common food products involved in the staphylococcal intoxication outbreaks with
S. aureus as the most common etiological agent in the dairy industry (
2, 3).
S. aureus can often be found as part of the normal microflora in humans and animals, but also on the hands, mouth, and feces in asymptomatic human carriers (
4, 5). The occurrence of staphylococcal infections is very common in dairy cows causing mastitis, which can be manifested in a subclinical form if it is not diagnosed early in the infection (
6). Produced enterotoxins in milk are resistant to the processes of pasteurization and canned sterilization (
7, 8).
Staphylococcus spp. is considered as one of the indicators of milk hygienic storage which can be affected by product manipulation and hence could indicate milk quality which is fundamental for cheese production (
9). The growth of
S. aureus and its SEs can occur mostly in raw milk as a result of inadequate temperature during storage, or in the first steps of cheese making, especially in raw milk cheese, when the replication of the enterotoxigenic strains is not prevented by the activity of lactic acid bacteria (
10). The cheese-making process alters
S. aureus activity and its capacity to resist various factors in the cheese matrix such as physicochemical characteristics and type of bacterial communities. Recontamination can happen during processing and can affect the final quality of the product, especially in maturated/ripened cheeses. It should be noted that SEs might not be detected with immunological assays due to lost serological activity but can still remain biologically active (
11). Despite food outbreaks, SEs can be involved in causing allergic reactions, multisystem disorders, or even lethal outcomes for specific population categories (
12, 13, 14).
The immunoenzymatic methods are used for the detection of SEs. The ELFA–Enzyme-linked Fluorescent Assay is incorporated in the VIDAS system. Molecular methods are used for the genetic identification of staphylococcal toxins. According to Priego et al. (
15), PCR and VIDAS are compatible and complementary for the detection of SEs.
Gene expression in microorganisms can be affected by various environmental conditions which can stimulate or inhibit the expression of genes responsible for toxin production. Therefore, gene expression in certain environmental conditions can be an indicator of microorganism toxicity.
The main objectives of this study were to detect the occurrence of
Staphylococcus aureus in different types of cheese in North Macedonia, to identify the incidence of the SEs genes (
sea,
seb,
sec,
sed,
see) by utilizing PCR, and to detect the SEs potential in detected strains by utilizing VIDAS SET 2.
MATERIAL AND METHODS
Enumeration, isolation and biochemical identificationIn the period of 15 months (from January 2018 to March 2019), 270 samples of different types of cheese (cow’s milk cheese and mixed cow-sheep milk cheese) were collected from 15 dairies grouped according to the country’s administrative regions: Polog n=5, Vardar n=3, Skopje n=2, Eastern n=2, Southeastern n=1, Pelagonia n=1, Southwestern n=1. The cheese samples from cooked milk were collected in the final product package at different periods of the ripening time (from 2 weeks to 5 months). The sterile initial dilutions were prepared and homogenized. Enumeration of coagulasepositive staphylococci was performed according to ISO 6888-1 “Horizontal method for enumeration of coagulase-positive staphylococci” (
16). Samples were cultured on to Baird Parker agar with Egg Yolk Tellurite Emulsion (Sigma, B1051) and the Petri dishes were incubated at 37 °C for 48 h. The typical colonies were confirmed with the coagulase test (Remel™ Coagulase Plasma, R21060) and were further identified by GP ID cards of the Vitek 2 automatic identification system (Biomerieux). A suspension with an optical density of 0.52-0.63 McFarland in 3 ml of 0.45% saline solution was prepared from each isolate and was checked with the Vitek Densicheck densimeter. Each suspension was connected with the corresponding GP card containing 64 biochemical tests, and the identification was finalized in 6-8 hours.
Detection of enterotoxigenic strainsThe ability of strains to produce SEA, SEB, SEC, SED, and SEE was assessed with the automated immune-enzymatic ELFA (Enzyme Link Fluorescent Assay) VIDAS SET 2 system (Biomerieux). The test can detect 7 types of enterotoxins (SEA, SEB, SEC1, SEC2, SEC3, SED and SEE). We did not have appropriate manufacturer instructions for strain testing, so we modified the manufacturer’s general protocol with SET RPLA and ENTEROTOX-F (Denka Seiken Co., Ltd., Japan) extraction strep (
17, 18). Pure colony culture was inoculated into a test tube with 5 ml Brain heart infusion broth (Oxoid, CM1135). After 24 hours of incubation at 37 °C, the broth was centrifuged at 4 °C for 15 minutes at 4,000 g. The pH of the supernatant was adjusted to 7.5-8.0 using NaOH 1N. 500μl were transferred to the first well of the VIDAS SET2 strip and the test was performed in 80 min.
A protocol for SE gene detection was established with multiplex PCR which was based on detecting pairs of primers for the sequences, according to the protocol by the European Reference Laboratory for coagulase-positive staphylococci (EU-RL-CPS, Anses, France). The following reference strains were used: FRI S6 (
sea, seb), FRI 326 (
see), FRI 361 (23s Rna,
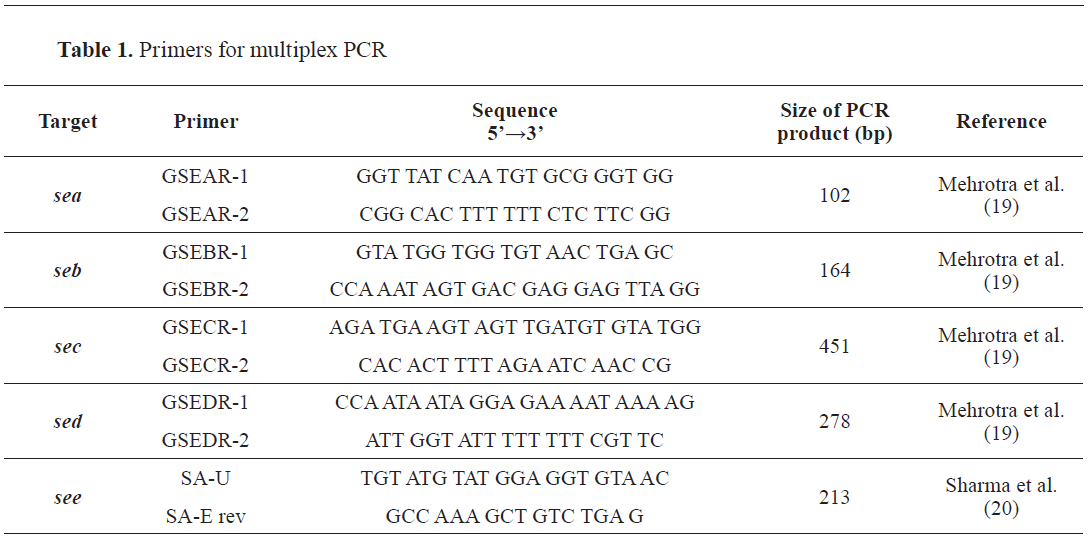
sec, sed, ser, seg, sei, sej), as positive controls obtained from EU-RL-CPS. The 5 pairs of primers designed for the specific target genes are given in
Table 1.
Bacterial DNA isolation was performed from fresh culture on a pure bacterial isolate from nonselective TSA agar by suspending several colonies in 50 μl Phosphate Buffered saline (PBS, Sigma Aldrich). The lysis was performed by incubating the suspension for 10 minutes at 95 ºC in MRC Thermo shaker 50 (UK), (
18). The resulting thermolysate was then centrifuged at 10,000 g/4 minutes.
The following protocol was used for the products in the master mix: Initial denaturation at 95 °C for 2 minutes, 35 cycles of: Denaturation (94 °C for 30 seconds); Adhesion of primers (60 °C for 30 seconds); Extension (72 °C for 60 seconds), and final extension at 72 °C for 10 minutes.
PCR products were separated by standard gel electrophoresis using 2% agarose gel (Agarose, Millipore) with 1xTBE buffer (Invitrogen, Thermo Fisher Scientific). Before pouring the gel, ethidium bromide was added (Sigma, Е1510) (3μl/100 ml gel). A marker with a known molecular mass of 100 bp DNA ladder (Solis Biodyne) containing standard fragments of 100 to 1,000 bp was used to estimate the size of the fragments obtained. PCR products were applied with a volume of 3 μl previously mixed with 1.5 μl per 3x Loading buffer (Thermo).
The electrophoresis was working at a voltage of 90 V and a duration of 90 min. After electrophoresis, the gels were visually read with a UV transilluminator (Gel Doc, XR + Bio-Rad).
Data analysisThe obtained results for the presence of enterotoxigenic potential that were obtained with 2 different methods were used to determine the diagnostic sensitivity and selectivity of the methods as well as positive and negative predictive values using a 2 × 2 contingency table. Diagnostic sensitivities and specificities were calculated as follows: 1. Sensitivity (%)=[True Positive/(True Positive+False Negative)]×100; 2. Specificity (%)=[True Negative/(True Negative+False Positive)]×100. The positive predictive value (PPV) (%)=[True Positive/(True Positive+False Positive)]×100. The negative predictive value (NPV) (%)=[True Negative/(True Negative+False Negative)]×100. (
21)
RESULTS
Out of 270 samples, 27 (10%) had CPS growth and 5 (1.8%) had coagulase-negative strains growth. The total viable counts of the CPS were in the range of 20 to 6,500 (1.3-3.8 log) cfu/g sample, which according to the legislation were deemed for further testing for the presence of SEs. All 27 isolates were positive on the coagulase test and were identified as
S. aureus with Vitek 2 GP-ID card with 95-99% accuracy. Out of 11 strains in 2018, 5 were positive for enterotoxin production detected with VIDAS, and in 2019 6 out of 16 samples or 11 strains (40.7%) from total samples were positive. The conventional PCR method has detected the marked genes in 7 (25.9%) strains, in 3 strains from 2018 , and in 4 strains from 2019. One of the strains from 2019 was negative on the VIDAS test but was positive for the presence of sea gene. Out of 7 strains that were positive for SE genes, n=3 (42.8%) had
seb, n=2 (28.6%) had
sea, and n=2 (28.6%) strains had the
sec gene. The percent of enterotoxogenic strains detected with the both methods per year are summerized in
Fig. 1, while the visualisation of the
seb and
sec genes with gel elecrophoresis is shown in
Fig. 2.
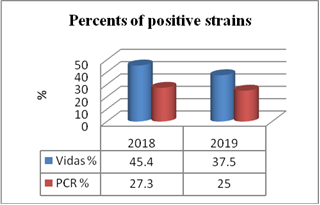 Figure 1.
Figure 1. Percent of positive strains with VIDAS system and PCR method
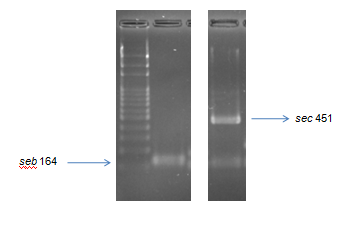 Figure 2.
Figure 2. Gel electrophoresis of multiplex PCR for detection of genes (bp)
Data statistics for the methods and the results are given in the
Table 2.
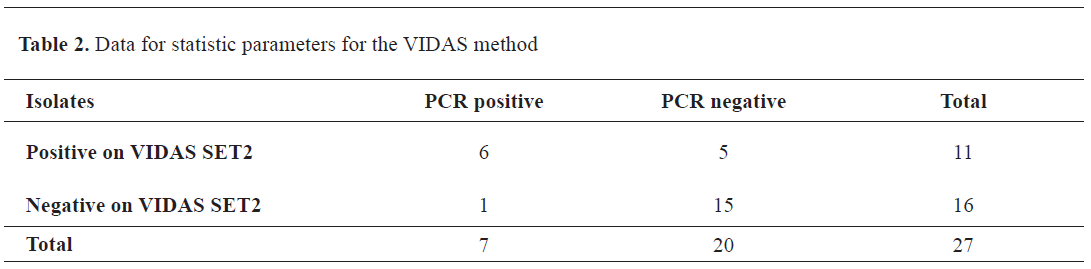
Considering that the PCR test detected the toxin genes and not their products, the data were used as a reference for calculating the VIDAS parameters: diagnostic sensitivity 85.7%, diagnostic specificity 75%, PPV=54.5% (true positives), and NPV=93.7% (true negatives).
DISCUSSION
According to the Book of rules for microbiological criteria (Off. G. of R.M. 100/2013), only samples showing counts of CPS higher than 105 cfu/g are further tested for the presence of enterotoxins (
22). All CPS counts were less than 105 cfu/g and were identified as
S. aureus as in the study of Medeiros et al. (2019) (23). Conversely, the study of Sampaio and Nader Filho (
24), showed that 50% of the samples in Brazilian cheeses (Mato Grosso) had CPS counts higher than the upper limit according to the local legislation. The occurrence of CPS varies ranging from 10% in our study, 20,48% in the study of Radovanovic et al. (
25) up to 80% (
23).
In the study of Vernozy-Rozand et al. (26), it was reported that 5.9% among 187 analyzed strains of
Staphylococcus spp. isolated from milk, whey and goat cheese were able to produce the SEs detected with VIDAS SET 2. In our previous study, 10.5% of the positive strains detected by the VIDAS SET 2 test were isolated from different types of cheese (
27). The current study findings (40.7%) are in compliance with the study of Holeckova et. al. (
28) which reported 39.2% SEs positive samples out of 51 isolated strains from sheep milk cheese produced in Slovakia. However, the sensitivity and specificity of the ELFA method may differ depending on the purity of the reagents and the level of the toxin expression. Aitichou et al. (
29) elaborated that the sensitivity limit, cross-reactivity, and interferences are affected by the analyzed matrix (alkaline phosphatase) and could be considered as significant disadvantages of the immunoassays. Hence, there is an ongoing need for further improvement of toxin detection methods (
30).
The presence of SEs in
S. aureus isolates is highly variable. In our study, SEs genes were found in 25.9% of
S. aureus strains, but there are reports with prevalence ranging from 3.6% (
31) to 100% (
32). The sec gene was detected in 26.8% in the current study which is similar to the findings of 21% (
23) and 25% (
33), but lower than those reported by Arcuri et al. (
31) 72.9%. In contrast, the
sea and
see genes were detected in 40% and
seb in 30% of analyzed Karich-cheese (
34), which is similar to our findings for the
seb gene (42.8%) and
sea gene (28.6%). The
see was not detected, whereas
sec gene was found in 28.6% of the samples. In another study
sea was detected in 4 (1.3%),
seb in 2 (0.6%), and
sed in 1 (0.3%) sheep cheese (
35).
The lack of correlation between the biochemical and molecular assays is already reported in the literature (
36). Jørgensen et al. (
36) and Kérouanton et al. (
37) suggested that the variations in primers sequences or presence of inhibitors in the annealing step of the primers could offer an explanation. Another possibility is that the PCR could not detect all genes present on one plasmid, like
sed, sej and
ser. This could be the reason for the positive detection of sed with VIDAS which did not present a band on the gel electrophoresis. Therefore, all samples showing VIDAS positive findings of enterotoxins should be tested for the presence of all possible combinations of enterotoxin genes by PCR. Also, the differences in the prevalence of different genes may depend on the country, local ecological origin of the strains, the sensitivity of the detection methods, sample number and the type of analyzed samples. Analysis of DNA sequences could solve the problem and reveal the actual discrepancies in the genes that could be present in the isolates.
CONCLUSION
All CPS counts of enterotoxigenic
S. aureus strains in the cheese samples were lower than the limit in the Book of rules for microbiological criteria. These results highlight the need for implementation of a National Monitoring Plan for the detection of enterotoxins and further evaluation of the legislation, especially reviewing the limit of CPS counts. Furthermore, there is a need of implementing higher hygienic practices and measures in all phases of production, processing, and distribution of dairy products, therefore guaranteeing high quality and safety. PCR method is an important tool to supply corresponding data about the genetic potential of the isolates. Immune-enzymatic assay is usually used to detect the presence of toxins in foods when the counts of
Staphylococcus aureus present were large enough to produce detectable enterotoxins. There is an ongoing need for further optimization of the existing analytical methods by achieving low or no interference with the food matrices, and by including newly described and known enterotoxins.
CONFLICT OF INTEREST
The authors declare that they have no potential conflict of interest with respect to the authorship and/or publication of this article.
ACKNOWLEDGMENTS
This research was supported by the Food institute at the Faculty of Veterinary Medicine – Skopje. The authors would like to express their gratitude to the colleagues Prof. Kiril Krstevski and Zaga Popovska for their scientific advices and technical support.
AUTHORS’ CONTRIBUTION
MRM conceived the study and wrote the manuscript. KB contributed with the practical performance of the study and supervised all the stages of the study. MP, KB, DJ and PS contributed to the study design. KB and PS gave critical revision and contributed to the final version and performance of the manuscript.

 10.2478/macvetrev-2022-0010
10.2478/macvetrev-2022-0010



