Abstract
Transmissible spongiform encephalopathies (TSEs) are a group of neurodegenerative diseases with a chronic and fatal course, which are caused by a misfolded form of the cellular prion protein that is encoded by the host. The purpose of this work was to evaluate the resistance and genetic susceptibility to prion diseases in Pelibuey sheep from Mexico. The sequences of 99 Pelibuey sheep from the central and southwestern regions of Mexico were analyzed to determine the polymorphisms related to resistance and genetic susceptibility to scrapie, the Hardy-Weinberg equilibrium test and the D-Tajima test was used to identify the effect of evolutionary forces on the PRNP gene. Twelve non-synonymous polymorphisms Q101R, M112T, A116P, G127A, A136V, M137T, L141F, H143R, R154H, Q171R, Q171H, N176K were detected, in addition two synonymous substitutions 231R (agg/cgg) and 237L (ctc/ctg) were found. As a result of the sequence analysis, the ARR allele was not under the Hardy-Weinberg equilibrium, indicating that there is an evolutionary force at work, and the D-Tajima suggest the existence of purifying genetic selection. In conclusion, Pelibuey sheep exhibit genotypes for the PRNP gene that make them resistant to developing classic scrapie; at least 98% of the population is a carrier of an allele related to scrapie resistance; while for atypical scrapie there is a very high probability that an outbreak occurred in the herds, since the ARQ allele in combination with the L substitution at codon 141 confers susceptibility to carrier sheep.
Keywords: PRNP gene, polymorphisms, genetic susceptibility, genetic selection, Pelibuey
INTRODUCTION
Scrapie is a deadly, neurodegenerative TSE disease that affects sheep and goats. TSEs are caused by infectious protein particles (prions), these molecules represent a misfolded form of the non-pathogenic cellular prion protein, encoded by the host, designated as PrPc. The gene that encodes the prion protein is known as
PRNP and the sequence of this gene is highly conserved in mammals (
1). In sheep, the prion protein gene is located on chromosome 13 and consists of 3 exons of 52, 98 and 4028 base pairs (bp), separated by 2 introns of 2421 and 14031 base pairs, the open reading frame it is in exon 3 and the resulting prion protein is 256 amino acids long (
2). Numerous polymorphisms of the DNA sequence of the PRNP gene have been described, most of them are within the open reading frame and several of them, especially at codons 136, 154 and 171 have been closely linked to disease susceptibility (
3). Sheep that are carriers of the PRNP genotype with VRQ and/or ARQ alleles are considered more susceptible to classic scrapie; while the PRNP genotypes that include the AHQ allele and the AF141RQ allele seem to confer greater susceptibility to atypical scrapie (
4). The ARR (alanine/arginine/arginine) allele is associated with resistance to natural and experimental infection with scrapie and bovine spongiform encephalopathy (BSE) (
5). The effect of other alleles is poorly understood and differs between populations. Polymorphisms in other codons, in particular substitutions of the amino acids Methionine (M) for Threonine (T) at codon 112 or Leucine (L) for Phenylalanine (F) at codon 141 (
6) seem to provide increased resistance to classic scrapie. Selective breeding programs, aiming to control classic scrapie, assume that the risk of acquiring the disease in sheep is determined by the genotype of the
PRNP gene (
7). This association between the
PRNP genotype and susceptibility to scrapie is the basis of breeding programs for resistance to scrapie in different countries. In 2001, the National Scrapie Scheme (NSP) was launched to assist sheep farming to reduce frequencies of susceptible genotypes in the United Kingdom (UK). The increasing frequency of the ARR/ARR genotype is considered an effective way to eradicate scrapie (
8), but the efficiency depends on several factors such as the size of the population, the frequency of the ARR allele and its relationship with economic performance (
9).
Several arguments have been raised regarding possible consequences of selection on
PRNP, including the risk of loss of genetic diversity. Breeding programs are criticized because they inevitably reduce the genetic variability available in a breed and the consequences are especially dangerous in breeds in terms of extinction (
10). There are reports that there is no significant relationship between
PRNP genotypes and litter size in sheep except for some breeds (
9). The results of the study by Brandsma et al. (
11) in Texel sheep indicated that selection of the ARR/ARR genotype had a small positive effect on litter size and a small negative effect on weight at 135 days. Likewise, the results of a study in the Ripollesa breed suggest that a favorable breeding program for the ARH haplotype could increase the litter size in sheep of this breed. In contrast, favorable selection for the ARR haplotype to increase genetic resistance to classical scrapie could reduce sheep prolificacy (
12). Few works have reported the association between the polymorphism of the
PRNP gene with postnatal survival. However, there is a general opinion that susceptible sheep outnumber resistant sheep and that the wild-type allele (ARQ) is associated with increased survival under harsh environmental conditions (
13). Furthermore, Gubbins and Cook reported associations between lamb survival and
PRNP genotype in the most common breeds of sheep in Great Britain (
14).
As part of the scrapie eradication programs, the selection of alleles that are related to greater resistance to the disease, as is the case of the ARR allele, could have a negative effect since the increase of this allele could result in a loss of genetic variability and a reduction in the effective size of sheep populations implying that genetic drift could increase as well as inbreeding (
15). The study of predisposition to scrapie is complicated because there are different
PRNP genotypes in different animals and breeds. There appears to be a relationship between breed and susceptibility to scrapie (
7). Despite the evidence that has been published on the global distribution of prion diseases, especially in sheep and goats as they are widely traded species, there is a concern that the disease may appear in countries where it has not been reported still, since this implies a high risk for the sheep and goat production industry. Therefore, the potential for resistance or susceptibility conferred by the genetic load of the herds should be evaluated in regions where there are no reports of the disease.
If the presence of polymorphisms related to genetic resistance to scrapie has contributed to the absence of this disease in sheep from Mexico, then sheep can be expected to carry alleles related to genetic resistance to scrapie. The objective of this study was to evaluate the resistance and genetic susceptibility to prion diseases in relation to the evolution of the
PRNP gene in Pelibuey type sheep from Mexico, through the analysis of genotypic and allelic frequencies. The Pelibuey sheep is a small to medium sized animal (between 35 and 80 kg), which instead of wool has hair, with three basic colors: brown, white and combined; on some occasions they can present areas of black hair. Although this breed has a low terminal efficiency in meat production, its biological importance lies in its ability to adapt to different climates and its ability to reproduce throughout the year (
16). The studies carried out to discover the polymorphisms in the resistance/susceptibility to scrapie disease allow us to see the richness of alleles and the genetic diversity that are important for the conservation of genetic resources.
MATERIAL AND METHODS
Sampling
Ninety-nine blood samples were obtained from
Pelibuey sheep from the central and southwestern regions of Mexico (27 and 72 samples, respectively), with an average age of 1 to 3 years. The blood samples were obtained by puncture of the jugular vein, collected in vacutainer tubes containing EDTA anticoagulant. They were transported to the laboratory using refrigerated coolers, in accordance with the guidelines of the terrestrial animal manual of the World Organization for Animal Health for the collection and shipment (
17).
Extraction of DNA Genomic DNA extraction was carried out with the Qiagen Dneasy kit using the protocol for DNA extraction by following the indications of the manufacturer.
Electrophoresis technique and determination of DNA concentration To check the integrity of the nucleic acids agarose gels of 1% were made, the samples stained with Ethidium bromide were placed and once the run was finished, the results were observed using the gel in a transilluminator with UV light. The concentration of the DNA was determined in a UV light spectrophotometer at 260 nm to measure the absorbance of an aliquot (1 μl) of the DNA obtained.
PRNP gene amplification PCR amplification of the
PRNP gene was performed in a final volume of 50 μl with 200 ng template DNA using the TopTaq Master Mix Kit (QUIAGEN) by following the indications of the manufacturer. The coding region was amplified using the following primers: F1 5´-ATGGTGAAAAGCCACATAGGCAGT-3´ and R1 5´-CTATCCTACTATGAGAAAAATGAG-3´ (
18). The amplification products were separated on a 1.5 % agarose gel, to verify the amplification product (771 base pairs).
DNA sequencing Sequence determination was performed on both strands of the PCR product. Samples were sequenced by Roslin Institute Neurobiology Division Laboratories in Edinburgh, Scotland and at the Transmissible Spongiform Encephalopathies Laboratory in Zaragoza, Spain. PCR fragments were sequenced with the Big Dye kit from Applied Biosystems.
The analysis sequencing was carried out with the DNA Sequencher 5.4.6 software, which made the readings of the sequences locating the order of the nucleotides, generated the chromatogram and prepared the report in Fasta format. The
Ovis aries Prion Protein sequence was taken from GenBank (accession number: 493887) to compare polymorphisms.
Statistical analysis The allele and genotype frequencies (%) were calculated as follows: F= (a/b) x 100, where
F – is the allele and genotype frequencies (%)
a - represents the number of times each allele or genotype was found
b - represents the total number of alleles or genotypes in the animals evaluated
The steady state of the population was determined according to Hardy-Weinberg (comparison of observed homozygotes and heterozygotes/expected homozygotes and heterozygotes) and the difference was analyzed using the chi-square statistical test. A statistical analysis of the sequences of the
PRNP gene of Pelibuey sheep was also carried out to look for signs of evolutionary forces on this gene for which the D-Tajima test was used. The analysis involved 198 nucleotide sequences and 840 positions in the final dataset. Evolutionary analyses were conducted in MEGA 7.
RESULTS
According to the analysis of the
PRNP gene sequence in Pelibuey sheep, twelve nonsynonymous polymorphisms: Q101R, M112T, A116P, G127A, A136V, M137T, L141F, H143R, R154H, Q171R, Q171H and N176K, in addition two synonymous substitutions 231R (agg/cgg) and 237L (ctc/ctg) were detected.
The analysis of the allelic frequencies in codons 136, 154 and 171, 190 alleles related to resistance(ARR; ARQ) were observed, representing 96%; while 5 AHQ alleles corresponding to 2.53% of the population have an allele whose function is unknown (
Table 1).
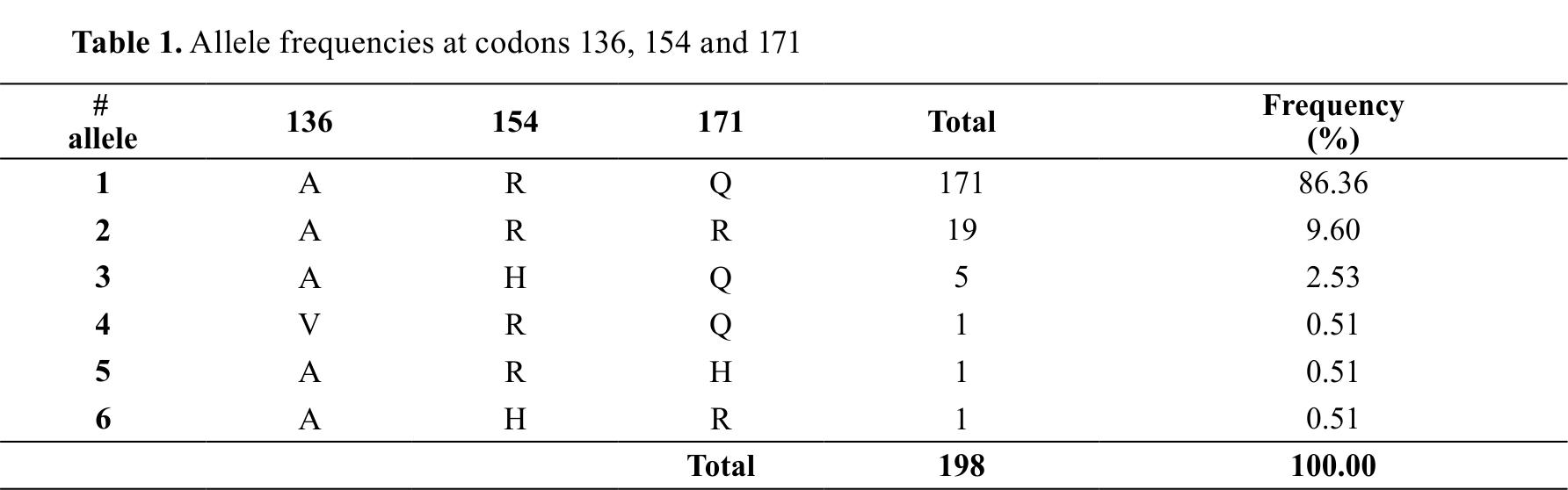
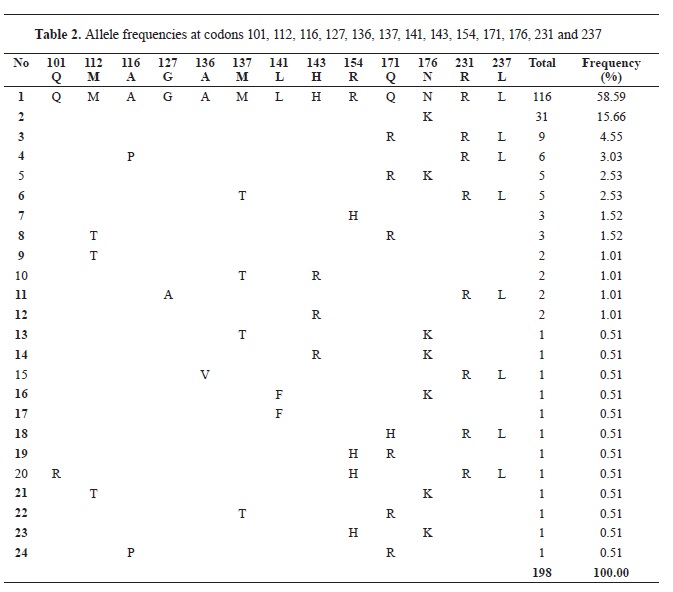
In the present study, polymorphic sites were detected in other codons of the
PRNP gene that have been previously reported. The Q101R polymorphism was only found with a frequency of 0.51%; the M112T with a frequency of 3.03%; A116P with 3.53%; G127A and L141F with 1.01%; M137T with 4.55%; H143R with 2.53% and N176K with 20.70%. The allele frequency for the silent substitutions 231R as well as for 237L was 71.21% (
Table 2).
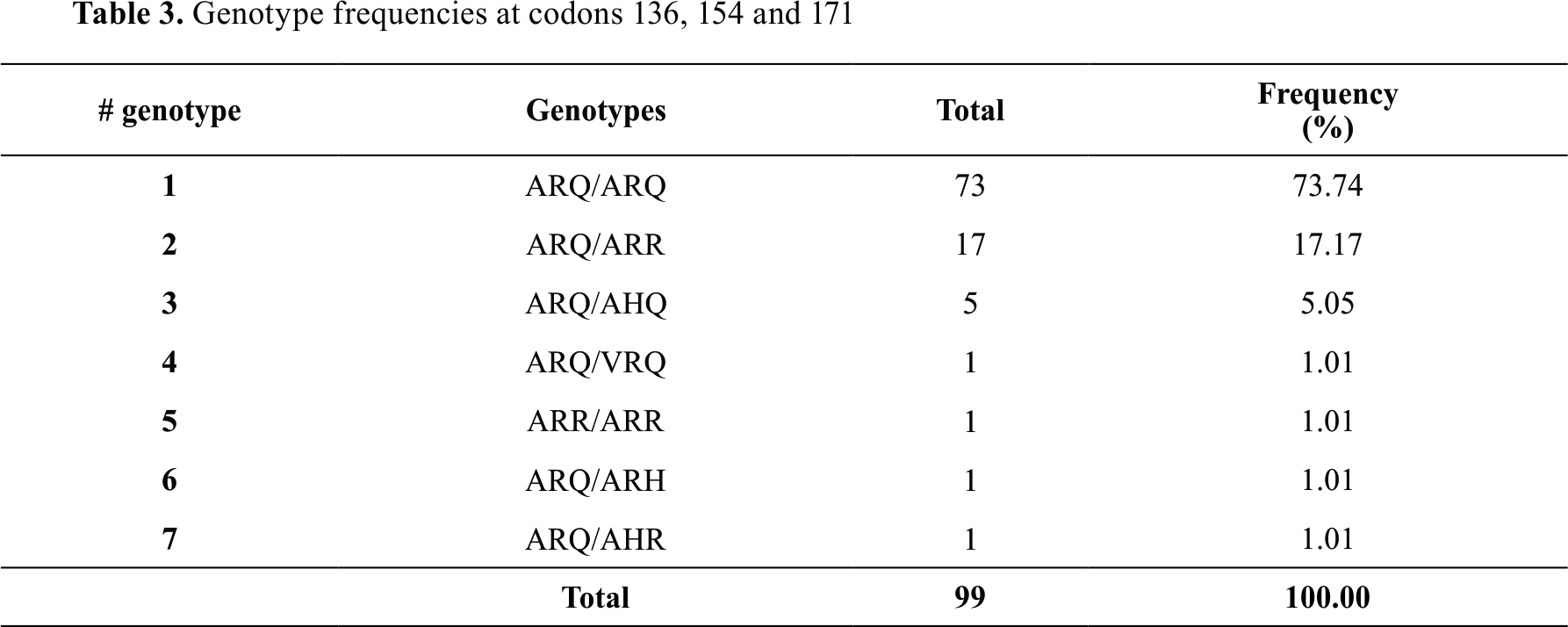
When analyzing the genotypes at positions 136, 154 and 171, the ARQ allele was found in 99% of the population (ARQ/ARQ, ARQ/ARR, ARQ/AHQ, ARQ/VRQ, ARQ/ARH and ARQ/AHR), while the VRQ allele related to the highest susceptibility was found in 1% (ARQ/VRQ) (
Table 3).
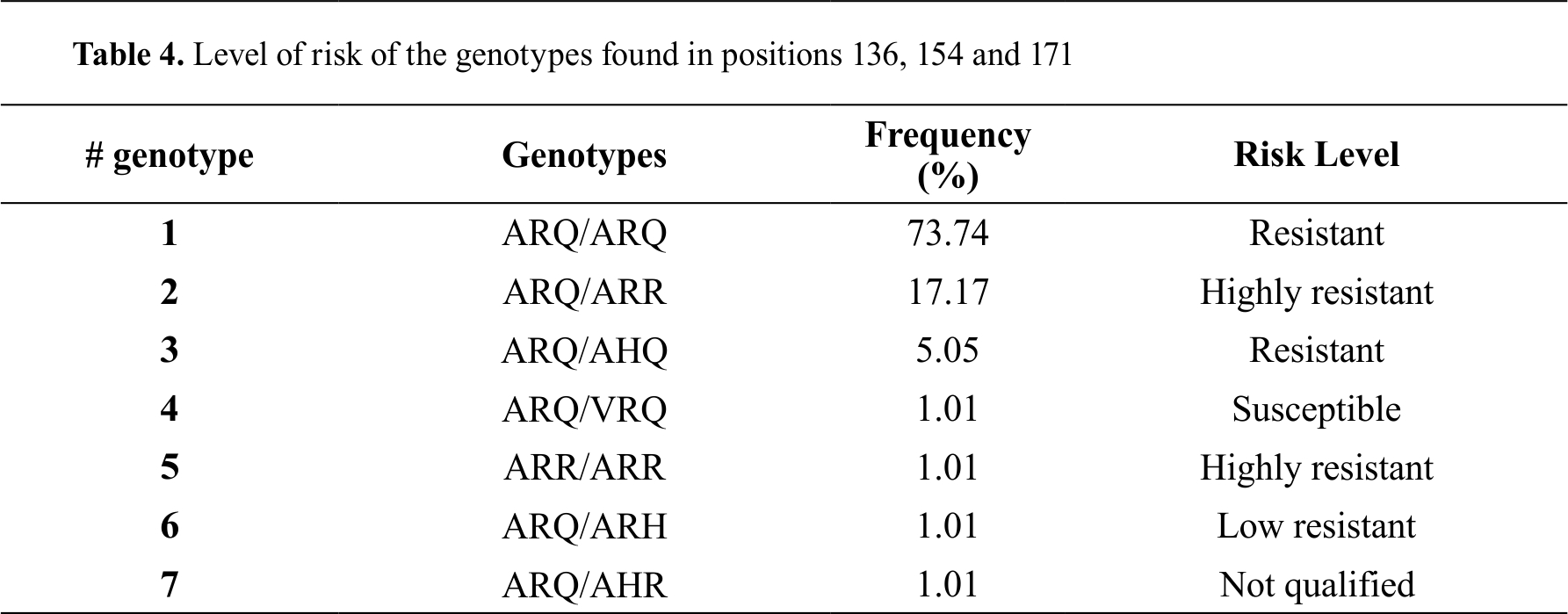
The analysis of the risk level of the genotypes found in positions 136, 154 and 171 was also carried out, according to the
PRNP genotype classification, based on the susceptibility to scrapie. 97.98% of the genotypes found in Pelibuey sheep it is classified as resistant to classic scrapie disease (
Table 4).
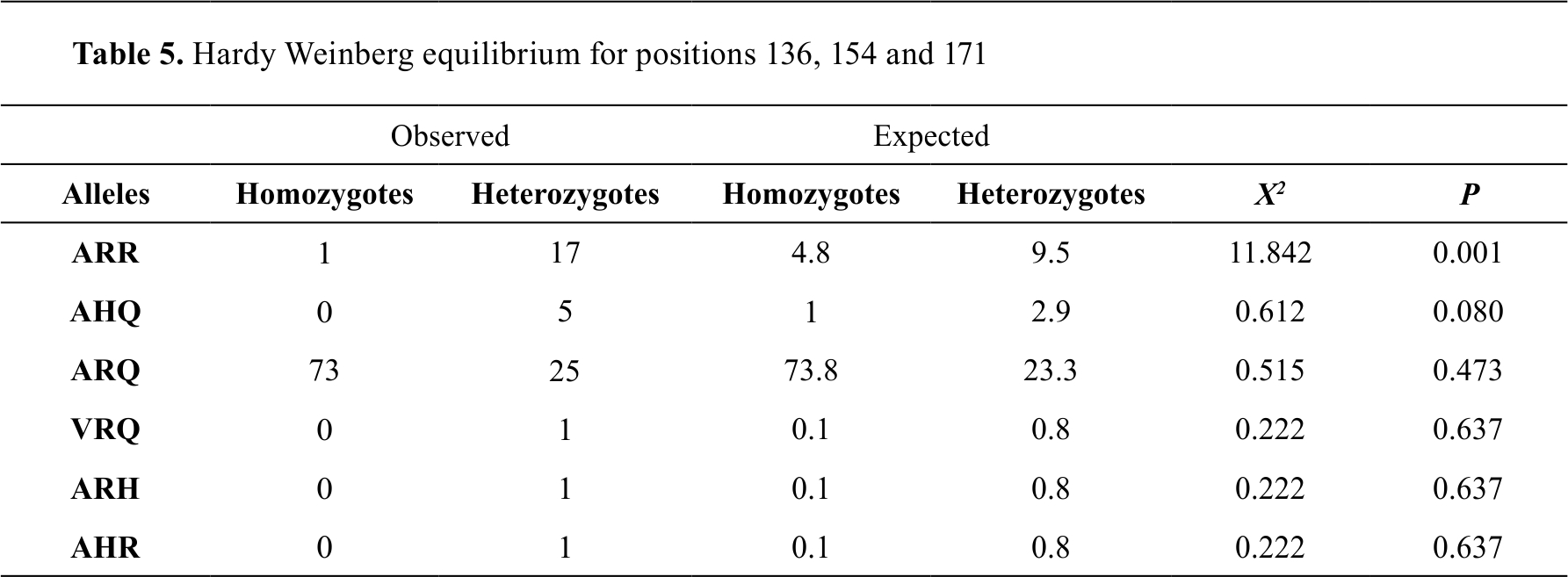
The Hardy-Weinberg equilibrium was used to calculate the number of homozygotes and heterozygotes that would be expected after a generation with random mating and the larger the increase in the differences between
the observed and expected values, the further away from Hardy-Weinberg equilibrium; therefore, the allele that was not under this balance was the ARR, which indicates that there is an evolutionary force that is acting on this allele (
Table 5).
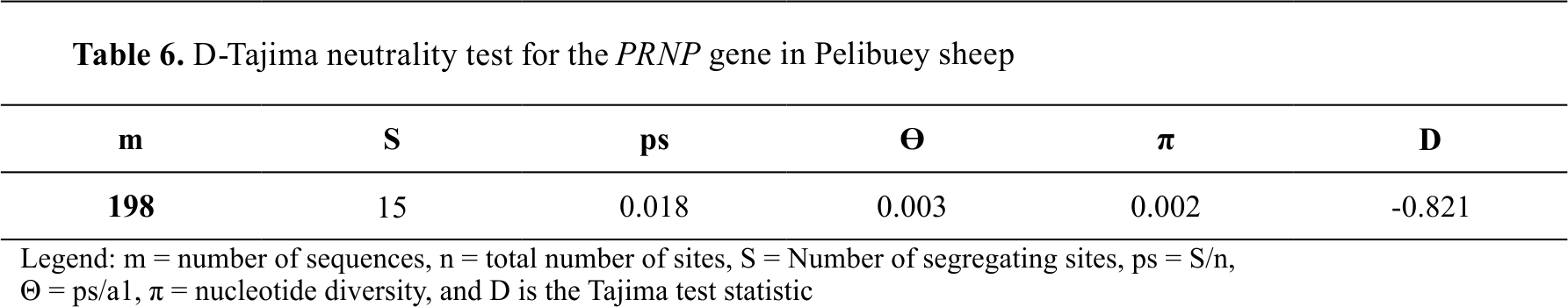
When the 198 nucleotide sequences were analyzed using the D-Tajima test, the value found in the
PRNP gene of Pelibuey sheep was -0.821 and seems to demonstrate the existence of purifying genetic selection (
Table 6).
DISCUSSION
There are no reports of
PRNP genotyping in Pelibuey sheep in Mexico, as in other countries where scrapie disease has not been reported. Since
PRNP genotyping is not a routine test in these countries, it can be suggested that there is a selection of individuals that have phenotypes that correspond to traits related to better production. In addition, the published works on Pelibuey sheep are associated with the improvement of the productive characteristics of these animals. Based on the hypothesis that the genetic composition of Pelibuey sheep influences the fact that scrapie has not been reported in herds in Mexico so far, the results of our study suggest that this proposal is correct, since 98.98 % of Pelibuey sheep have at least one allele carrying resistance to classic scrapie.
In the analysis of the
PRNP gene sequence in Pelibuey sheep carried out in this work, twelve non-synonymous polymorphisms and in two synonymous substitutions were detected. All nonsynonymous substitutions have been reported in countries with scrapie except for the Q101R substitution. The M112T substitution was reported in Great Britain (
6). A116P has been observed in the United States in the St. Croix breed (
19). G127A has been reported in the United States (
20). M137T was reported in British sheep (
21) and in Italy in the Sardinian breed (
22). L141F has been reported in British sheep (
21) and in Finland it was found in the Landrace sheep breed (
23). H143R was reported in sheep from Oklahoma (
24). N176K has been reported in Italy in the Sardinian breed (
22). These studies were carried out in countries where the disease has been present for several years and correspond to commercial breeds or commercial crossbreeds with local breeds. In them, the allelic frequencies were higher than those found for Pelibuey sheep in our study.
Non-synonymous substitutions that have been reported in non-scrape countries are Q101R, M112T, A136V, M137T, L141F, H143R, R154H, Q171R, Q171H, and N176K. The Q101R substitution has been observed in regions of China (
25). M112T has been reported in China (
9) and in regions of Turkey it has been found in indigenous breeds (
26). M137T was confirmed in China in sheep of the Hu breed (
9) and in regions of Turkey (
26). L141F occurs in sheep of Turkish breeds (
26) and in China (
25). H143R is observed in regions of Turkey (
26) and an analysis on 16 local breeds is reported in China (
25). It is important to mention that these investigations were carried out in native breeds. Allele frequencies reported in countries free of scrapie are like those found in Pelibuey sheep from Mexico. The synonymous substitutions 231R and 237L that were identified in Pelibuey sheep have been reported in native Turkish sheep breeds (
27) and native Ethiopian sheep breeds (
28). Considering that in Mexico as well as in Ethiopia and some regions of Turkey the presence of prion disease has not been reported and that these synonymous substitutions is found in practically two thirds of the Pelibuey sheep that were evaluated in the present study, it is suggested that these 2 variations may influence resistance and/or susceptibility to scrapie.
Several studies have revealed that the ovine
PRNP gene is highly polymorphic. However, only some polymorphisms at positions 136, 154 and 171 have been clearly associated with the occurrence of scrapie. The most frequently found variants in different breeds around the world for these three polymorphisms are ARR, ARQ, AHQ and VRQ (
29). All these
PRNP variants were found in Pelibuey sheep. All of the six alleles found in this work have already been reported previously. ARQ has been found in Italian sheep breeds (
22); native sheep from Bulgaria (
30), in China (
9); in Turkey (
31) and in Brazil, (
32). VRQ, which is associated with the highest susceptibility to scrapie, has been observed in herds of the Italian Sarda breed (
22); in native European sheep breeds (
33); in Turkey (
31) and in Brazil the absence of the allele is reported in some breeds (
32). The frequency for the VRQ allele found in Pelibuey breed herds is consistent with the frequencies in countries where the disease is absent, since there is no artificial selection based on this locus. ARR, which is associated with the highest resistance, has been found in Italy for sheep of the Sardinian breed (
22), in other European breeds (
33); in China sheep of the Hu breed (
9) and in Bulgaria (
30). Such high frequencies in countries with scrapie are because in these places they have already implemented the selection of animals carrying this allele. Even though in Pelibuey sheep the frequency of the ARR allele is less than 10%, possibly due to the use of homozygotes carrying other alleles, the possible strategies to increase resistance to scrapie should not necessarily consist in the selection of animals carrying the ARR, but also to determine the possible associations with other polymorphic sites within the
PRNP genome that confer resistance not only to classic scrapie, but to possible atypical forms of the disease, as well.
The results obtained from the analysis of synonymous and non-synonymous substitutions in the
PRNP gene in Pelibuey sheep suggest that this gene has been subjected to purifying selection and not to a balanced selection as reported by Slate (
34) for sheep of Norwegian origin. This difference is possibly due to the fact the disease has not been reported in Mexico, while Norway is a country where scrapie exists. This suggests that scrapie may be a factor contributing to the selection of alleles related to genetic susceptibility to this disease. Other factors can cause patterns of purifying selection: an excess of rare synonymous variants in a population may indicate recent population expansion, followed by insufficient time to establish the balance between the occurrence of new mutations and their loss through genetic drift (
35). Regarding this, in Mexico there are quantitative records of sheep populations in different periods (
36), observing that the herds constantly suffer population expansions, nevertheless, it is difficult to know for sure the level of genetic exchange and the time spent in the herd, due to the lack of records of the sheep that enter and leave the productive units. In addition, it is to be expected that individuals with phenotypes corresponding to traits related to better production are being selected. Purifying selection has been reported for this same gene and its paralogous genes in some studies: the first was a sequence analysis in 26 mammalian species (
37); the second was an evaluation of the
PRNP gene in bovines and the results indicate that a strong purifying selection acted on this gene (
38) and the third consisted in the exploration of the
PRND gene that is paralogous to the
PRNP gene and whose evolution has been by virtue of purifying selection among mammalian taxa (
39). Sezgin et al reported that a complex selection favoring excessive replacement changes together with weak purifying selection possibly driven by frequency-dependent selection is driving
PRNP sequence evolution (
40).
Since scrapie is considered the prototype of prion diseases, the determination of polymorphisms provides a key point for understanding why some countries continue to be free of scrapie even though they are carriers of alleles considered high susceptibility. According to the results and their comparison with previously published articles, there are three aspects to emphasize. First, indigenous breeds of sheep appear to exhibit a higher level of resistance to prion diseases compared to commercial breeds. Second, the analysis of the
PRNP gene allows elucidating the resistance and genetic susceptibility of the herd. Third, allely frequencies exhibited by Pelibuey sheep in our work, suggest that there is a resistance to classic scrapie and susceptibility to atypical scrapie, with the possibility that an outbreak may occur, since the ARQ allele in combination with the L substitution at codon 141 confers susceptibility to carrier sheep.
For future research, the racial effect should be investigated for the understanding of the intraspecific barrier of the prion protein and for the understanding of the interspecific barrier of
PRNP, the polymorphisms that are shared by several species should be studied, such is the case of positions 141 and 143. Likewise, due to the level of resistance observed in this animal group and the absence of scrapie disease in Mexico, it is important to evaluate all breeds of sheep and goats found in the territory to create a risk map for prion diseases. In addition, it is necessary to carry out a retrospective study in the history of sheep that are currently considered resistant to scrapie to find some evidence of a possible evolutionary force that could have been exerted by some epizootics of scrapie in the past.
CONCLUSION
Pelibuey sheep exhibit genotypes for the PRNP gene that make them resistant to developing classic scrapie; at least 98% of the population carries an allele related to scrapie resistance; therefore, in the near future, the genetic composition of these animals could allow the absence of cases of the disease in Mexico; while for atypical scrapie there is a very high probability that an outbreak occurred in the herds. The natural selection that seems to have governed the evolution of the PRNP gene in Pelibuey sheep is the purifying type selection, since in an attempt by human beings to select the optimal productive and reproductive characteristics of their herds, the alleles that grant an intermediate protection for the disease.
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no potential conflict of interest with respect to the authorship and/or publication of this article.
ACKNOWLEDGMENTS
This research was supported by Faculty of Veterinary Medicine and Zootechnics, Meritorious Autonomous University in Puebla, Mexico and the Faculty’s Academic Body of Emerging Diseases, Bioinformatics and Molecular Modeling.
AUTHORS’ CONTRIBUTIONS
FRA, and AVC conceived and designed the study, supervised the experiments and data analysis, interpreted the results, performed a major part of the experiments and wrote the manuscript. FRA, CFPR, RAB, RSMM and JAGD participated in designing the study, and contributed to finalizing the manuscript.
References
1. O’Neill, G.T., Donnelly, K., Marshall, E., Cairns, D., Goldmann, W., Hunter, N. (2003). Characterization of ovine PrP gene promoter activity in N2a neuroblastoma and ovine foetal brain cell lines. J Anim Breed Genet. 120, 14-123. https://doi.org/10.1046/j.1439-0388.2003.00381.x
2. Goldmann, W., Baylis, M., Chihota, C., Stevenson, E., Hunter, N. (2005). Frequencies of PrP gene haplotypes in British sheep flocks and the implications for breeding programmes. J Appl Microbiol. 98, 1294-1302.
https://doi.org/10.1111/j.1365-2672.2005.02568.x PMid:15916643
5. Paludi, D., Thellung, S., Chiovitti, K., Corsaro, A., Villa, V., Russo, C., et al. (2007). Different structural stability and toxicity of PrP(ARR) and PrP(ARQ) sheep prion protein variants. J Neurochem. 103, 2291- 300.
https://doi.org/10.1111/j.1471-4159.2007.04934.x PMid:17919292
6. Saunders, G.C., Lantier, I., Cawthraw, S., Berthon, P., Moore, S.J., Arnold, M.E., et al. (2009). Protective effect of the T112 PrP variant in sheep challenged with bovine spongiform encephalopathy. J Gen Virol. 90, 2569-574.
https://doi.org/10.1099/vir.0.012724-0 PMid:19587133
7. McIntyre, K.M., Trewby, H., Gubbins, S., Baylis, M. (2010). The impact of sheep breed on the risk of classical scrapie. Epidemiol Infect. 138, 384-392.
https://doi.org/10.1017/S0950268809990537 PMid:19678970
8. Drogemuller, C., Leeb, T., Distl, O. (2001). PrP genotype frequencies in German breeding sheep and the potential to breed for resistance to scrapie. Vet Rec. 149, 349-352.
https://doi.org/10.1136/vr.149.12.349 PMid:11594380
9. Guan, F., Pan, L., Li J., Tang, H., Zhu, C., Shi, G. (2011). Polymorphisms of the prion protein gene and their effects on litter size and risk evaluation for scrapie in Chinese Hu sheep. Virus Genes. 43, 147-152.
https://doi.org/10.1007/s11262-011-0609-5 PMid:21556743 PMCid:PMC3124648
10. Sartore, S., Rasero, R., Colussi, S., Acutis, P.L., Peletto, S., Soglia, D., et al. (2013). Effect of selection for scrapie resistance on genetic diversity in a rare and locally adapted sheep breed: The case of Sambucana. Livest Sci. 157, 75-80.
https://doi.org/10.1016/j.livsci.2013.08.006 11. Brandsma, J.H., Janss, L.L.G., Visscher, A.H. (2005). Association between PrP genotypes and performance traits in an experimental Dutch Texel herd. Livest Prod Sci. 95, 89-94.
https://doi.org/10.1016/j.livprodsci.2004.12.011 12. Casellas, J., Caja, G., Bach, R., Francino, O., Piedrafita, J. (2007). Association analyses between the prion protein locus and reproductive and lamb weight traits in Ripollesa sheep. J Anim Sci. 85, 592-597.
https://doi.org/10.2527/jas.2006-308 PMid:17060422
13. Woolhouse, M.E., Coen, P., Matthews, L., Foster, J.D., Elsen, J.M., Lewis, R.M., et al. (2001). A centuries-long epidemic of scrapie in British sheep? Trends Microbiol. 9, 67-70.
https://doi.org/10.1016/S0966-842X(00)01912-0 PMid:11173245
14. Gubbins, S., Cook, C.J., Hyder, K., Boulton, K., Davis, C., Thomas, E., et al. (2009). Associations between lamb survival and prion protein genotype: analysis of data for ten sheep breeds in Great Britain. BMC Vet Res. 5, 3.
https://doi.org/10.1186/1746-6148-5-3 PMid:19159456 PMCid:PMC2637852
15. Alfonso, L., Parada, A., Legarra, A., Ugarte, E., Arana, A. (2006). The effects of selective breeding against scrapie susceptibility on the genetic variability of the Latxa Black-Faced sheep breed. Genet Sel Evol. 38, 495-511.
https://doi.org/10.1186/1297-9686-38-5-495 PMid:16954042 PMCid:PMC2689259
16. Aguilar-Martínez, C.U., Berruecos-Villalobos, J.M., Espinoza-Gutiérrez, B., Segura-Correa, J.C., Valencia-Méndez, J., Roldán-Roldán, A. (2017). Origen, historia y situacion actual de la oveja pelibuey en Mexico. Tropical and Subtropical Agroecosystems 20, 429-439.
17. OIE (2005) Sampling methods. OIE Manual on Terrestrial Animals. 3-15.
18. Acin, C., Martin-Burriel, I., Goldmann, W., Lyahyai, J., Monzon, M., Bolea, R., et al. (2004). Prion protein gene polymorphisms in healthy and scrapie-affected Spanish sheep. J Gen Virol. 85, 2103-2110.
https://doi.org/10.1099/vir.0.80047-0 PMid:15218196
19. Seabury, C.M., Derr, J.N. (2003). Identification of a novel ovine PrP polymorphism and scrapie-resistant genotypes for St. Croix White and a related composite breed. Cytogenet Genome Res. 102, 85-88.
https://doi.org/10.1159/000075730 PMid:14970684
20. Heaton, Leymaster, K.A., Freking, B.A., Hawk, D.A., Smith, T.L.P., Keele, J.W., et al. (2003). Prion gene sequence variation within diverse groups of U.S. sheep, beef cattle, and deer. Mamm Genome. 14, 765-777.
https://doi.org/10.1007/s00335-003-2283-y PMid:14722726
21. Saunders, G.C., Cawthraw, S., Mountjoy, S.J., Hope, J., Windl, O. (2006). PrP genotypes of atypical scrapie cases in Great Britain. J Gen Virol. 87, 3141-3149.
https://doi.org/10.1099/vir.0.81779-0 PMid:17030846
22. Vaccari, G., Scavia, G., Sala, M., Cosseddu, G., Chiappini, B., Conte, M., et al. (2009). Protective effect of the AT137RQ and ARQK176 PrP allele against classical scrapie in Sarda breed sheep. Vet Res. 40, 19.
https://doi.org/10.1051/vetres/2009002 PMid:19171116 PMCid:PMC2695041
23. Hautaniemi, M., Tapiovaara, H., Korpenfelt, S.L., Sihvonen, L. (2012). Genotyping and surveillance for scrapie in Finnish sheep. BMC Vet Res. 8, 122.
https://doi.org/10.1186/1746-6148-8-122 PMid:22831168 PMCid:PMC3414783
24. DeSilva U., Guo X., Kupfer D.M., Fernando S.C., Pillai A.T., Najar F.Z., et al. (2003). Allelic variants of ovine prion protein gene (PRNP) in Oklahoma sheep. Cytogenet Genome Res. 102, 89-94.
https://doi.org/10.1159/000075731 PMid:14970685
25. Lan, Li J., Sun, C., Liu, Y., Zhao, Y., Chi, T., et al. (2014). Allelic variants of PRNP in 16 Chinese local sheep breeds. Arch Virol. 159, 2141-2144.
https://doi.org/10.1007/s00705-014-2048-9 PMid:24643335
26. Alvarez, Gutierrez-Gil, B., Uzun, M., Primitivo, F.S., Arranz, J.J. (2011). Genetic variability in the prion protein gene in five indigenous Turkish sheep breeds. Small Rumin Res. 99, 93-98.
https://doi.org/10.1016/j.smallrumres.2011.03.043 27. Ün, C., Oztabak, K., Ozdemir, N., Akıs, I., Mengi, A. (2008). Genotyping of PrP gene in native Turkish sheep breeds. Small Rumin Res. 74, 260-264.
https://doi.org/10.1016/j.smallrumres.2007.06.002 28. Teferedegn, E.Y., Yaman, Y., Un, C. (2020). Five novel PRNP gene polymorphisms and their potential effect on Scrapie susceptibility in three native Ethiopian sheep breeds. BMC Vet Res. 16, 122.
https://doi.org/10.1186/s12917-020-02336-0 PMid:32349749 PMCid:PMC7189463
29. Hunter, N., Moore, L., Hosie, B.D., Dingwall, W.S., Greig, A. (1997). Association between natural scrapie and PrP genotype in a flock of Suffolk sheep in Scotland. Vet Rec. 140, 59-63.
https://doi.org/10.1136/vr.140.3.59 PMid:9023905
30. Sirakov, I., Peshev, R., Christova, L. (2011). Genetic predisposition of some Bulgarian sheep breeds to the scrapie disease. Virus Genes. 43, 153-159.
https://doi.org/10.1007/s11262-011-0615-7 PMid:21533749
31. Meydan Yuceer, B., Degirmenci, R., Ozkan, M.M., Yildiz, M.A. (2012). Prion protein gene polymorphism and genetic risk evaluation for scrapie in all Turkish native sheep breeds. Virus Genes. 45, 169-175.
https://doi.org/10.1007/s11262-012-0744-7 PMid:22528641
32. Ianella, P., McManus, C.M., Caetano, A.R., Paiva, S.R. (2012). PRNP haplotype and genotype frequencies in Brazilian sheep: issues for conservation and breeding programs. Res Vet Sci. 93, 219-225.
https://doi.org/10.1016/j.rvsc.2011.06.025 PMid:21816449
33. Lühken, G., Lipsky, S., Peter, C., Erhardt, G. (2008). Prion protein polymorphisms in autochthonous European sheep breeds in respect to scrapie eradication in affected flocks. Small Rumin Res. 75, 43-47.
https://doi.org/10.1016/j.smallrumres.2007.07.010 34. Slate, J. (2005). Molecular evolution of the sheep prion protein gene. Proc Biol Sci. 272, 2371-2377. https://doi.org/10.1098/rspb.2005.3259 PMid:16243700 PMCid:PMC1559970
35. Glatt, C.E., DeYoung, J.A., Delgado, S., Service, S.K., Giacomini, K.M., Edwards, R.H., et al. (2001). Screening a large reference sample to identify very low frequency sequence variants: comparisons between two genes. Nat Genet. 27, 435-438.
https://doi.org/10.1038/86948 PMid:11279528
36. SIAP (2021). Sheep, Livestock population, 2012-2021. https://www.gob.mx/cms/uploads/attachment/file/744954/Inventario_2021_ovino.pdf
37. van Rheede, T., Smolenaars ,M.M., Madsen, O., de Jong, W.W. (2003). Molecular evolution of the mammalian prion protein. Mol Biol Evol. 20, 111-121.
https://doi.org/10.1093/molbev/msg014 PMid:12519913
38. Seabury, C.M., Honeycutt, R.L., Rooney, A.P., Halbert, N.D., Derr, J.N. (2004). Prion protein gene (PRNP) variants and evidence for strong purifying selection in functionally important regions of bovine exon 3. Proc Natl Acad Sci U S A. 101, 15142-15147.
https://doi.org/10.1073/pnas.0406403101 PMid:15477588 PMCid:PMC524052
39. Tsangaras, K., Kolokotronis, S.O., Ulrich, R.G., Morand, S., Michaux, J., Greenwood, A.D. (2014). Negative purifying selection drives prion and doppel protein evolution. J Mol Evol. 79, 12-20.
https://doi.org/10.1007/s00239-014-9632-1 PMid:25038839
40. Sezgin, E., Teferedegn, E.Y., Un, C., Yaman, Y. (2022). Excessive replacement changes drive evolution of global sheep prion protein (PRNP) sequences. Heredity (Edinb). 128, 377-385.
https://doi.org/10.1038/s41437-022-00520-6 PMid:35273383

 10.2478/macvetrev-2022-0030
10.2478/macvetrev-2022-0030





