Bartonella species are recognized as important zoonotic pathogens, affecting a wide range of animals worldwide, including ruminants, canids, felids, rodents, bats, reptiles, and humans (
1). The genus
Bartonella includes many bacterial species, such as
B.
henselae, the causative agent of human cat-scratch disease (CSD) (
1, 2).
B.
henselae is a Gram-negative, facultative, intracellular bacteria associated with an expanding spectrum of clinical signs in its natural and accidental hosts. The domestic cat (
Felis catus) is considered as the main reservoir. The pathogen is transmitted by vectors (bloodsucking arthropods), particularly fleas (
2). In addition to many documented hosts, an increasing number of vectors, including ticks, have been suspected or confirmed to be associated with the transmission of
Bartonella spp. (
3, 4). Infected cats are typically asymptomatic but can spread the infection to humans through cutaneous trauma, primarily by scratches and bites (
5). The clinical signs of the disease are present in both immunocompromised and immunocompetent individuals (
6). Laboratory diagnosis of
Bartonella infections is based on direct detection and identification of the pathogen using classical microbiological methods (bacterial culturing) and molecular diagnostic methods (PCR, Real-Time PCR, Sequencing), as well as indirect diagnosis using serological methods for detection of specific antibodies against
Bartonella spp. (immunofluorescence assay and ELISA) (
1, 6).
The first report of cat-scratch disease in the Republic of North Macedonia (R.N. Macedonia) was described in 2020 in a female patient presenting clinical symptoms of mild fever and axillary lymphadenopathy in whom the diagnosis of CSD was made based on the detection of IgM and IgG antibodies against
Bartonella henselae in the serum, using an indirect immunofluorescence assay (IFA) (
7).
There are no studies or published data on the prevalence or the detection of
B.
henselae in cats in R.N. Macedonia. Here we describe the first case of
B.
henselae infection in a cat from R. N. Macedonia, confirmed through molecular testing, following the suspicion of a CSD in a seven-year-old boy.
MATERIAL AND METHODSCase background and samplingA blood sample of a physically healthy feral cat that was in contact with a seven-year-old boy with symptoms of CSD was collected in a K2-EDTA tube from a local veterinarian and delivered to the Laboratory of Microbiology at the Faculty of Veterinary Medicine-Skopje for analysis.
CultureThe blood was stored overnight at -18 °C to lyse the erythrocytes. After thawing, 200 μl of blood were inoculated in duplicate on blood agar (BA) plates, supplemented with 5% defibrinated sheep blood. The plates were rotated, allowing the blood to distribute on the agar surface without mechanical spreading and incubated agar side down at 37 °C in an 8% CO₂ atmosphere. Following incubation for 3 days, the plates were inverted agar side up and inspected daily. Suspect colonies started appearing on the ninth day following inoculation.
DNA extractionА 200 μl suspension of a single bacterial colony (isolate MKD1) in Phosphate Buffered Saline (PBS) was prepared and boiled for 30 minutes. After centrifugation (12,000 X g for 10 minutes), the supernatant was transferred into a new sample tube, and the DNA extraction was performed on a SaMag-24 Automatic Nucleic Acids Extraction System (Sacace Biotechnologies, Italy), using the Bacterial DNA Extraction Kit (Sacace Biotechnologies, Italy), following the manufacturer’s protocol. The evaluation of the extracted DNA (quality and quantity) was done using the NanoDrop 2000c UV-Vis Spectrophotometer (Thermo Scientific, USA).
Molecular identificationTo identify the bacterial isolate, two distinct regions in the bacterial genome were amplified using previously published conventional PCR protocols (
8, 9, 10), with adapted master mix preparation following the recommendations of the amplification kit manufacturer.
In the first protocol (
8), a partial sequence of the bacterial 16S ribosomal RNA (rRNA) gene was amplified for universal bacterial identification using the primers presented in
Table 1 (PCRs 1-3). Briefly, three separate reaction mixtures were prepared in a final volume of 25 μl, containing 12.5 μl of 2x AmpliTaq Gold 360 Master Mix (Applied Biosystems), 1 μl of each, forward and reverse primers (final concentration 0.4 μM), 7.5 μl of nuclease-free water, and 3 μl of extracted DNA. The amplification was performed on a SimpliAmp Thermal Cycler (Applied Biosystems) using the following thermal protocol: initial denaturation at 95 °C for 10 min followed by 30 cycles consisting of denaturation at 94 °C for 30 sec, annealing at 60 °C for 1.5 min, extension at 72 °C for 2 min. The amplification was finalized with an additional extension step at 72 °C for 7 min.
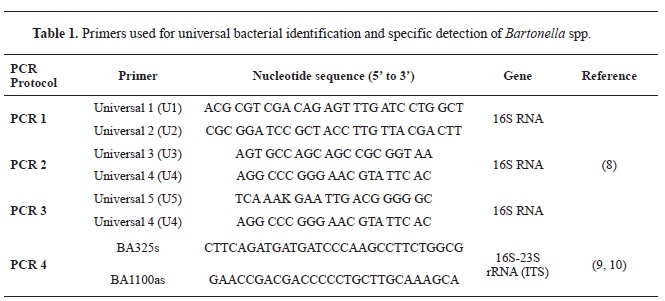
The second protocol, used for specific detection of
Bartonella spp., amplified a partial sequence (420-780 bp) of the 16S–23S rRNA Gene Intergenic Spacer Region (ITS) using the primers presented in
Table 1 (PCR 4) (
9, 10). Briefly, the reaction mixture was prepared the same way as for the 16S rRNA protocol, with the only difference being the final concentration of the primers (1 μM). The amplification was performed on a SimpliAmp Thermal Cycler (Applied Biosystems) using the following thermal protocol: initial denaturation at 94 °C for 10 min followed by 40 cycles consisting of denaturation at 94 °C for 30 sec, annealing at 66 °C for 30 sec, extension at 72 °C for 50 sec.
The PCR products were visualised by electrophoresis on a 1.5% agarose gel stained with GelRedTM (Biotium, USA) (
9).
The PCR products were purified using the Wizard SV Gel and Clean Up System (Promega, USA), according to the manufacturer’s recommendations. Bidirectional sequencing, using the same primers used in all four PCRs, was performed on a SeqStudio Genetic Analyzer (Applied Biosystems, Singapore) using the BigDye v.1.1 cycle sequencing kit (Applied Biosystems, CA, USA). The obtained sequences were analysed and assembled using the Staden Package (version 2.0.0b11) (
11), and the identification was performed using the Basic Local Alignment Search Tool (BLAST) of the National Center for Biotechnology Information (NCBI) (
12).
The sequences from both the 16S ribosomal RNA (rRNA) gene and 16S–23S rRNA Gene Intergenic Spacer Region (ITS) were deposited in the GenBank and can be accessed under the accession numbers OQ120562 and OQ110535, respectively.
RESULTSAfter nine days of incubation, colonies typical for
Bartonella spp. were observed on the blood agar plates. The colonies appeared white-creamy, small and rough in texture. The characteristic colony morphology provided preliminary evidence for the presence of
B.
henselae.
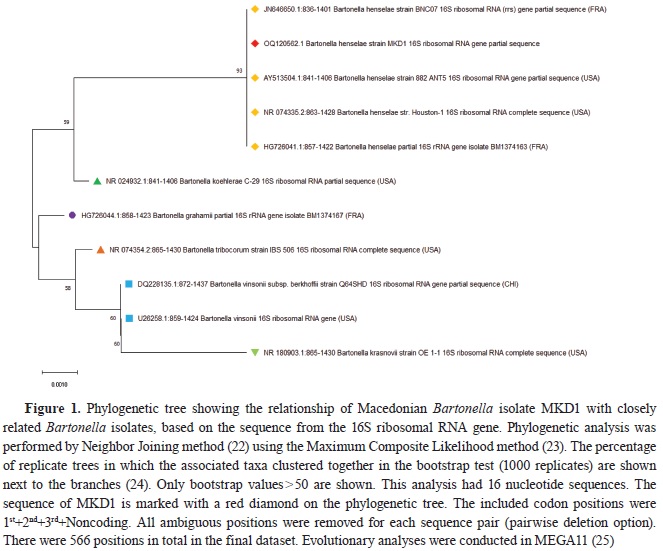

All three PCRs targeting the 16S ribosomal RNA (rRNA) gene (PCR 1-3,
Table 1) gave positive results for amplification of the specific DNA fragment, visualized as a fluorescent band on the agarose gel. The sequences from all three PCR products were combined, and a final consensus sequence of 566 bp in length was reached and compared with the publicly available data stored in the GenBank (NCBI). The comparison revealed high sequence homology with the sequences from the bacteria of the genus
Bartonella (>98.94%). Macedonian
Bartonella isolate (MKD1) clustered together with the isolates of
B.
henselae, sharing a 100% sequence identity, and formed a separate group from other
Bartonella species on the phylogenetic tree, supported by high bootstrap value (
Fig. 1).
The
Bartonella spp. specific PCR, targeting the 16S–23S rRNA Gene Intergenic Spacer Region (ITS), yielded a PCR product of approximately 600 bp. A consensus sequence of 565 bp was obtained and compared against the sequences deposited in the GenBank. Phylogenetic analysis revealed 100% identity with the sequences of the
B.
henselae, clustering together on a separate branch and forming a distinct group, supported by high bootstrap value (
Fig. 2).
DISCUSSIONThis study reports the first
B.
henselae infection in a cat in North Macedonia. The cat was asymptomatic, and the infection was confirmed by culture and molecular analysis. A blood sample from the suspected cat was cultured on a blood agar plate, and bacterial DNA was extracted from colonies resembling
B.
henselae. The sequences obtained for both the 16S rRNA gene and 16S – 23S rRNA (ITS) region showed high discriminatory power clustering the MKD1 isolate together with the isolates of
B.
henselae with 100% sequence identity, thus proving the PCR protocols used suitable for identification and differentiation of
Bartonella species.
The primary mode of
B.
henselae transmission among cats is through the flea
Ctenocephalides felis and their feces, where bacteria can remain viable for at least nine days (
13). Furthermore, ticks have also been recognised as potential vectors for
Bartonella transmission (
4).
B.
henselae DNA has been found in several species of ticks, including
Ixodes pacificus, Ixodes ricinus and
Rhipicephalus sanguineus (
4). Furthermore,
B.
henselae has been isolated from blood cultures or detected by PCR from individuals who experienced a tick bite; this reinforces the hypothesis that ticks might serve as vectors for
Bartonella spp. (
4, 10). Despite the potential low epidemiological significance, further investigation is required to understand vectors for
B.
henselae better (
10). This would underline the extensive range of arthropod hosts for
Bartonella spp.
Bartonella infections can persist in cats and fleas for a long time, and there is a positive association between flea infestation, lack of ectoparasiticide use and
B. henselae infection in cats (
14). The cat from our study was feral, thus we assumed it may have had a flea infestation. Outdoor access and living in multi-cat households present risk factors significantly associated with
B.
henselae infections in cats (
6, 13, 14).
Bartonella infections in cats and other animals have been reported worldwide. The seroprevalence rates of
Bartonella infections in cats in Europe vary significantly, with higher rates observed in European Mediterranean countries characterised by favourable conditions for flea infestation (
14). These rates range from 0% in Norway to as high as 71.4% in Spain (
5, 13, 14). The variance in seroprevalence observed in studies from Spain is likely attributable to the source of cats, as stray and shelter cats appear to be at greater risk due to potentially high flea exposure and lack of preventive measures (
5). The study of Alejandra Alvarez-Fernandez et al. (
5) confirms what is becoming increasingly apparent – cats living in the Mediterranean region seem to have higher
Bartonella spp. prevalence than other parts of Europe.
The occurrence of feline bartonellosis cases confirmed through PCR testing has been infrequent. Studies have shown that the prevalence of
Bartonella infection in cats, as determined through molecular methods, varies from 0% in northeast Germany to 83.5% in Italy (
13). These variations may be attributed to several factors, including geographical location, environmental conditions and flea exposure (
5, 13).
Regarding the fact that this is first report, the prevalence of the infection among the cat population in North Macedonia has yet to be determined. Seroprevalence is higher in cats over the age of one (
15), so future studies should focus on older cats. While the diagnostics and detection of
B.
henselae are primarily focused on stray cats, it is important to note that indoor cats and non-reservoir hosts like dogs can also pose relevant health concerns (
5, 13, 16). Cats infected with
B.
henselae, whether pets or strays, may pose a potential risk of infection to humans due to frequent and direct contact among them. Moreover, the clinical manifestations associated with this pathogen in human patients can be potentially affected by various factors such as a person’s immune system, variations in the strain’s virulence and co-infection with other pathogens (
13). It has been suggested that
B.
henselae infection is more likely to occur in younger individuals (
6) exposed to cats infested with fleas, such as the infected boy that was in contact with the cat from this study. This is supported by the case study of a young boy who was infected after prior contact with a cat (
6). Testing for
B.
henselae should be performed in healthy cats with a history of flea infestation, outdoor access and in pets owned by immunocompromised people (
14).
Isolation of
B.
henselae is a diagnostic challenge because of low bacteraemia and fastidious growth characteristics, with colonies usually visible 10-56 days after inoculation (
13, 17). The blood agar culture method described in this study effectively detects and isolates
B.
henselae. The extended incubation period of nine days allowed the slow-growing
Bartonella spp. to grow and form characteristic colonies. The colony morphology observed on the blood agar plates was consistent with previous reports of
B.
henselae colonies (
6, 18). Our findings also confirm that primary
Bartonella colonies can be obtained before the tenth day in blood culture and correspond with previous reports (
6, 18). Direct blood culture confirms the presence of bacteria, and it is preferable to serological tests as most outdoor cats, although not bacteremic, develop anti-
Bartonella antibodies (
6, 19).
Without a gold standard test, the definitive diagnosis of feline bartonellosis remains a challenge (
13). Serological testing alone is not recommended since there are false-positive test results, and it should be combined with cultural or molecular diagnostic methods, which are infrequently reported (
13, 20). To facilitate the growth of
Bartonella spp., an enrichment medium known as
Bartonella alphaproteobacterium growth medium (BAPGM) was specifically designed (
21). Furthermore, combining molecular and liquid culture methods increases the chances of
B.
henselae DNA detection in cats (
20). To detect the pathogen and enhance sensitivity and specificity, we have also used a combination of primary culture and molecular confirmation of the isolate.
The sequence obtained with the PCRs targeting the 16S rRNA gene showed 100% homology with
B.
henselae strains from France (JN646650 and HG726041) and the USA (AY513504 and NR074335) (
Fig. 1), while the 16S–23S rRNA ITS sequences showed 100% homology with the strains from Brazil (MT095053, MT095050), New Zealand (MF196158) and Germany (CP072898) (
Fig. 2). The high sequence homology observed between the MKD1 isolate and
B.
henselae isolates from different geographic regions reflects the stability in the targeted segments of the genome, making it a suitable tool for molecular identification, but not as a tool for molecular epidemiology.
CONCLUSIONOur results indicate that
B.
henselae infection in cats is present in North Macedonia. The blood agar culture method described in this study offers a reliable tool for detecting and isolating
B.
henselae from blood samples. The study confirms the identity of MKD1 as
B.
henselae, based on high sequence homology, clustering with known
B.
henselae isolates, and distinct grouping on phylogenetic trees.
There is limited information about
Bartonella infections in our country in humans and animals. Therefore, it is essential to increase awareness about the potential zoonotic risk of
B.
henselae infections among cat owners and the human population and investigate the prevalence of the disease among them. Cats without preventive flea treatment are significantly more likely to be infected, suggesting that vector-control programs are essential preventative measures and crucial to avoid
Bartonella infection.
CONFLICT OF INTERESTThe authors declare that they have no potential conflict of interest with respect to the authorship and/or publication of this article.
ACKNOWLEDGMENTSThis research was supported by the Faculty of Veterinary Medicine-Skopje.
AUTHORS’ CONTRIBUTIONIC conceived the study, supervised the bacteriological testing, interpreted the results and monitored manuscript writing. IS and AC performed the bacteriological testing, interpretation of the results and wrote the manuscript.AJ and LR were included in the bacteriological testing. ID, KK and ZPH performed the molecular study and participated in writing of the manuscript. All authors have revised and approved the final version of the manuscript.

 10.2478/macvetrev-2023-0028
10.2478/macvetrev-2023-0028


