Over the years several studies in Albania reported the presence of Salmonellosis in poultry, while the information on swine Salmonellosis is limited. A recent case in Albania reports
S. enterica serovar Choleraesuis var. Kunzerdof isolated in piglets from intensive and backyard farms during a septicemia form of infection (
6). The domestic pig population in Albania fluctuated substantially in recent years with a downward trend, and in 2021 over 160 thousand pigs were registered. More than 93.8% of pig farms are small-sized, mostly fatteners with less than 100 animals, 4.3% are breeder fatteners with less than 1,000, and only 10 farms (1.7%) are large commercial farms with more than 1,000 animals. Pig production is concentrated in the western and central parts of the country (
7).
The general clinical forms of Salmonellosis in swine are enterocolitis caused mainly by
S. enterica serovar Choleraesuis,
S. typhimurium, and septicemia which is due to
S. enterica serovar Choleraesuis. This last form is more problematic because it affects multiple organ systems (
8). Outbreaks caused by
S. enterica serovar Choleraesuis are often linked to “stress” conditions. The majority of affected pigs are weaned and younger than five months. Infection of pigs with
S. enterica serovar Choleraesuis occurs via oral route and leads to the colonization of lymphoid tissues in the digestive system. A septicemic infection caused by
S. enterica serovar Choleraesuis can be fatal even without warning symptoms. Postmortem examination reveals interstitial pneumonia, mesenteric lymphadenopathy, splenomegaly, hepatomegaly, and colitis. Identification and isolation of the organism are necessary for a conclusive diagnosis. Through direct culture,
S. enterica serovar Choleraesuis can be isolated from untreated septicemic pigs (
9).
The key factor that accelerates the spread of infectious pathogens is human activity and its impact. Numerous practices, such as industrial animal husbandry and the use of antibiotic growth promoters, have selected novel variations, such as hyper virulent and antimicrobial resistant clones (
10). Using antibiotics to prevent and treat bacterial diseases is a widely used practice in veterinary medicine. Furthermore, the inadequate application of antimicrobials is a major factor in the development of bacteria resistant to antibiotics in animals, which can then be passed on to humans via the food chain (
11, 12).
Salmonella spp. has been shown in numerous investigations to be resistant to antimicrobial drugs from various antibiotic groups. One reliable and affordable way to monitor antimicrobial resistance is through the multiple antibiotic resistances (MAR) index (
13, 14). It is a quick and simple method (
15). When a bacteria isolate comes from a source where antibiotics are used extensively and/or in significant quantities, the MAR index is more than 0.2 (
16).
The present study aimed to identify the serovar Choleraesuis during
Salmonella outbreaks in domestic pigs in central-western Albania, and to assess the antimicrobial resistance of circulating serotypes.
MATERIAL AND METHODSOrgan samples (lung, spleen, liver, kidney, and intestine) from 240 dead domestic pigs were collected by farm veterinarians and were sent to the laboratory. The ages of the animals ranged from 3 to 5 months. Samples were taken aseptically in sterile polyethylene sachets, and they were kept cold inside an insulated box with ice packs for the lab’s transportation. When the samples arrived at the lab, they were immediately processed for bacterial isolation.
Bacterial isolation and identificationThe primary culture and isolation was performed according to the Manual of diagnostic techniques of World Organization of Animal Health (WOAH 2018). The tissues from organ samples were homogenized in a small amount of sterile saline. A volume of 10 ml of homogenate was combined with 100 ml of non-selective enrichment broth, i.e. Buffered Peptone Water (Biolife, Milan, Italy). The cultured samples were incubated at 37 °C. After 24 hours, they were subcultured on both selective and non-selective agar i.e. Blood agar and MacConkey agar, and were incubated at 37 °C for 18-24 h in aerobic conditions. After incubation for 24±2 h, the samples were observed for colonial growth on Petri dishes. All presumptive colonies of
Salmonella were subcultured onto Xylose-Lysine- Deoxycholate (XLD) agar, Hektoen Enteric (HE) agar (Biolife, Milan, Italy), and were incubated at 37 °C for 24 h. Suspected colonies were tested biochemically using Triple Sugar Iron (TSI) and anti-
Salmonella (A-E+Vi) sera (Sifin diagnostics gmbh) on slide agglutination test according to the Kauffmann-White scheme (Supplement 2008-2010, no. 48). Further presumptive identification was performed automatically by Matrix-Assisted Laser Desorption–Ionization Time-of-Flight Mass Spectrometry, MALDI TOF Biotype Sirius (Bruker Daltonik GmbH, Germany).
Antimicrobial susceptibility testThe Kirby-Bauer disc diffusion method on Mueller-Hinton agar (MHA) (Biolife, Milan, Italy), was used to assess the antibiotic susceptibility of
Salmonella isolates in accordance with the guidelines provided by the Clinical Laboratory Standards Institute (CLSI 2018) and EUCAST disc diffusion methods (Version 3.0, April 2013). Approximately 3-4 colonies were picked to make a suspension of the organism to the density of a McFarland 0.5 turbidity standard. The inoculum was spread over the entire surface of MHA plate by swabbing uniformly in three directions. The antibiotic discs (Biomaxima S.A, Lublin, Poland) with a specified concentration were aseptically applied and incubated for 18 to 24 h at 37 °C. Antibiotics were chosen from 7 different classes, i.e. tetracyclines (doxycycline, oxytetracycline), penicillin (ampicillin, amoxicillin, cefalexin), aminoglycosides (gentamicin, spiramycin), fluoroquinolones (enrofloxacin, ciprofloxacin, ofloxacin), amphenicols (florfenicol), sulfonamides (sulfamethoxazole, trimethoprim), and polymyxin group (colistin) according to European Committee on Antimicrobial Susceptibility Testing (EUCAST). Based on recommendations released by CLSI 2018, the diameter of the zone of inhibition surrounding each disc was measured, recorded, and interpreted as resistance (R), sensitive (S), or intermediate (I). The formula MAR =
a/b was used to estimate multiple antibiotic resistance (MAR), where a is the number of antibiotics to which the test isolate was resistant and b is the total number of medications tested. Multi-drug resistance (MDR), extensively drug-resistant (XDR), and Pan Drug Resistance PDR were calculated.
RESULTSThe results of this study indicated that a 10% of organ samples (24/240) submitted in the laboratory were contaminated with
Salmonella spp., and among them, 16.6% (4 samples) were identified as
Salmonella enterica serovar Choleraesuis. Antimicrobial susceptibility tests of the 4
Salmonella isolates against 13 antimicrobial agents are presented in
Table 1. The percentage of
Salmonella resistance was 75.0% for ampicillin, amoxicillin, oxytetracycline, sulfamethoxazole, and trimethoprim followed by 50.0% for doxycycline, cefalexin, and spiramycin. the resistance to colistin was less frequently observed, only in 25.0% of the isolates. On the other hand, the isolates were sensitive to enrofloxacin, ciprofloxacin, and florfenicol in 100% followed by ofloxacin -75% sensitive and 25% intermediate.
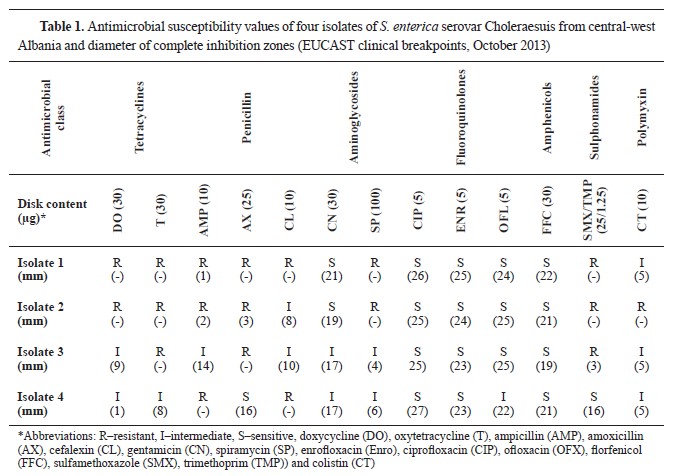
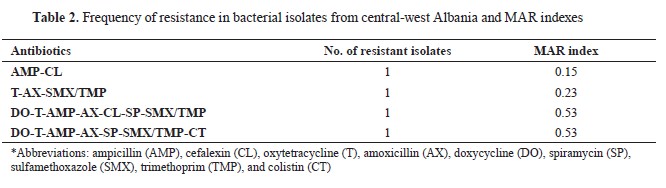

The frequency of antimicrobial resistance in
Salmonella related to the MAR index is shown in
Table 2. The results indicate that all
S. enterica serovar Choleraesuis isolates showed resistance to at least two antimicrobials, and 3 of them exhibited MDR. The resistance to oxytetracycline, amoxicillin, sulfamethoxazole, and trimethoprim was with a MAR index of 0.23. Ampicillin (75%) was present in almost all multi‐resistance patterns. Two isolates were observed to display resistance to seven antimicrobials with a MAR index of 0.53 (
Table 2).
The
Salmonella isolates from tissues and organs in the current study demonstrated multidrugresistant
S. enterica serovar Choleraesuis contamination. The test for antibiotic sensitivity employing fourteen distinct antibiotics revealed the presence of multidrug-resistant serovars.
Table 3 shows the antimicrobial resistance of the
Salmonella isolates. Three
Salmonella isolates showed MDR (Isolate 1, 2, 3). Specifically, isolate No.1 was resistant to at least one antibiotic in 4 antimicrobial categories (tetracyclines, penicillin, aminoglycosides, and sulphonamides); isolate No. 2 was resistant to at least one antibiotic in 5 antimicrobial categories (tetracyclines, penicillin, aminoglycosides, sulphonamides, and polymyxin), and isolate No. 3 was resistant to at least one antibiotic in 3 antimicrobials class categories (tetracyclines, penicillin, and sulphonamides). One isolate (No. 2) was resistant to 2 antimicrobial categories, and it was categorized as Extensive Drug Resistance. None of the isolates displayed Pan Drag Resistance or Non-Multi Drag Resistance.
DISCUSSIONMeat and meat products are among the basic products of the consumer’s basket. Albania, as a developing country, is behind the average consumption per capita compared to the developed countries. The meat processing industry has had a steady growth, although food safety remains an issue (
7). Special attention and care are paid to the safe and infectious-disease-free purchase of animals, biological material, and food. Salmonellosis continues to be a serious issue and financial burden for pork producers because of its presence and the risk for transmission to humans by consuming contaminated pork products.
S. enterica serovar Choleraesuis is the primary cause of paratyphoid fever in swine. Previously, it was reported that the experimental oral infection of
S. enterica serovar Choleraesuis in pigs resulted in interstitial pneumonia, septicemia, multifocal necrosis in the liver, massive ulceration, and necrotic colitis (
17). Confirmation of the disease requires bacterial isolation and identification. Undetected and asymptomatic infections in weaned piglets are most likely triggered by stress which lowers the immune system. The initial selection of antibacterial drugs is based on the sensitivity of
S. enterica serovar Choleraesuis isolates in a given area and by antimicrobial sensitivity testing.
The isolates of
S. enterica serovar Choleraesuis exhibit a range of resistance against several antimicrobial classes. The highest antimicrobial resistance is registered for the sulphonamide, penicillin, and tetracycline groups. This may be related to the routine use of single or combined antibiotics on the farms in order to prevent or treat various diseases. All isolates were susceptible to fluoroquinolones and florfenicol (100%). After analyzing the antibiotic susceptibility, 3 out of 4 isolates showed MDR, 1 isolate XDR, and none was categorized as a PDR pathogen. Findings from several authors (
18) state that in order to guarantee the proper implementation of these definitions, bacterial isolates should be tested against all or most of the antimicrobial agents in each category. The results should not be obscured or reported selectively. Combined antimicrobials can work more quickly and efficiently. Values of the MAR index >0.2 are related to the presence of bacteria in sources that are highly contaminated and with frequent use of antibiotics, whereas values <0.2 indicate bacteria from sources with low frequency of antimicrobial use (
19). When MAR index values are high, alert monitoring and corrective measures are necessary.
Other authors report similar results for several groups of antimicrobials, in cases of
S. enterica serovar Choleraesuis strains isolation with significant resistance to sulfonamide, tetracycline, beta lactam, and aminoglycoside, and frequent use in feed supplements in intensive farming of pigs (
20, 21, 22). Our results show that gentamicin (CN) from the aminoglycoside group still remains effective even though it has long been used for treatment of moderate to severe gram-negative infections in pigs. On the other hand, resistance to clinical antimicrobials such as trimethoprim, cephalosporins, and quinolone were far less common and frequently restricted to the human strains (
23).
These results present serious hazards to public health because these antibiotic classes are currently medications of choice for treating human Salmonellosis infections. Although
Salmonella’s antibiotic resistance and prevalence in the swine production chain has been extensively studied, still, several aspects related to molecular mechanism remains unknown (
24). Whole genome sequencing is now a common and helpful method for risk assessment, not used in Albania yet.
CONCLUSIONFarm-raised pigs in central-west Albania had Salmonellosis caused by
S. enterica serovar Choleraesuis. Therefore, pig farmers must implement monitoring, prevention, control, and eradication programs. Additionally, this study revealed that there is a significant danger to food safety due to the high frequency of multi drug resistant
Salmonella. This suggests a responsible use of antimicrobials by veterinarians, and appropriate antimicrobial management plans in affected farms.
CONFLICT OF INTERESTSThe authors declare that they have no known conflict of interest in the conduction of the current study.
ACKNOWLEDGMENTSThe authors would like to acknowledge the Faculty of Veterinary Medicine of Tirana and BIO-V Laboratory for providing intellectual contribution, and infrastructure in this study.
AUTHORS’ CONTRIBUTIONLL had the main idea, supervised laboratory work and data analysis, and wrote the manuscript. GJD followed up on the practical work and data analyses. AÇ made the sample collection and data collection. LT critically reviewed the manuscript.

 10.2478/macvetrev-2024-0023
10.2478/macvetrev-2024-0023


