The emergence of antimicrobial resistance (AMR) in Staphylococcus spp. in companion animals is a cause for concern and poses a significant threat to public health (1). Staphylococcus infections are commonly diagnosed in small animal clinical practice with most dog isolates showing resistance to at least one antibiotic (2). In addition, the emergence and spread of methicillin-resistant strains has led to the development of multidrug-resistant (MDR) bacteria (3), defined as resistance to at least one drug from three or more antibiotic classes (3, 4). These MDR strains exhibit resistance to almost all antimicrobials approved for veterinary use, posing a significant challenge in small animal practice. In 2021, EFSA identified S. pseudintermedius as one of the top three antimicrobial-resistant bacteria in the EU that poses a risk to the health of dogs and cats (5). Although S. seudintermedius is a commensal bacterium, as an opportunistic pathogen, it can be responsible for many infections, including skin infections, otitis externa, urinary, respiratory, and reproductive infections (5). In addition to S. pseudintermedius , other clinically important Staphylococci can also be isolated and cause infections, such as Staphylococcus aureus, Staphylococcus schleiferi, Staphylococcus intermedius, Staphylococcus hemolyticus and Staphylococcus hyicus (2, 4, 6) .
Methicillin resistance (MR) is one of the most important public health concerns in human medicine due to the high prevalence of methicillin-resistant (MRSA) infections that are now increasingly observed in isolates from dogs (10). In addition, methicillin-resistant S. pseudintermedius (MRSP) has been identified as one of the most important bacterial pathogens for companion animals in the European Union and is sporadically associated with human infections (11, 12, 13). Staphylococcus species acquire methicillin resistance mainly through the mec A gene, which encodes an altered penicillin-binding protein (PBP2a). The altered protein has a lower affinity for beta-lactam antibiotics, making them ineffective against methicillin-resistant strains. Methicillin-resistant strains of Staphylococcus Staphylococcus aureus spp. commonly exhibit resistance to oxacillin or cefoxitin, which serve as phenotypic indicators of methicillin resistance (7). While the mecA gene is the gold standard for MR identification, other mechanisms such as altered penicillin-binding proteins, β-lactamase hyperproduction, or the presence of less common methicillin-resistance genes may be responsible for the observed oxacillin/cefoxitin resistance in these isolates (8). However, another gene, mecC, has also been identified and contributes to this resistance. The mecC gene codes for an alternative penicillin-binding protein called PBP2c, which, like PBP2a produced by mecA, has a low affinity for beta-lactam antibiotics. In addition, the blaZ gene, which encodes for beta-lactamase production, is also crucial as it contributes to resistance against beta-lactam antibiotics and further complicates treatment options.
While research on AMR in companion animals has been extensive in Western Europe, including established monitoring systems and stewardship guidelines, studies and systematic data collection from the Balkan region remains limited (9, 10). In particular, data on the prevalence of AMR in Staphylococcus species isolated from dogs in North Macedonia is scarce. A single study by Cvetkovikj et al. (11) reported a significant prevalence of MRSP and MRSA among canine isolates in the country, highlighting the need for further investigation. To address the gap, this study aimed to evaluate phenotypic resistance profiles and examine the presence of key resistance genes (mecA, mecC, and blaZ) in 170 Staphylococcus species isolates obtained from canine clinical samples. The findings will contribute to the regional data on AMR trends and support the development of effective antimicrobial stewardship strategies.
MATERIALS AND METHODS
Strain collection A total of 170 Staphylococcus isolates from clinical samples from 170 dogs were analyzed over a five-year period (2019-2024). All samples were collected by private veterinarians and submitted to the laboratory for bacteriological diagnosis according to the clinical manifestations observed. Any data on previous antimicrobial treatment, breed, age, or sex were not available. The research was conducted as part of the Project FVMS-IPR-4, "Antimicrobial resistance in bacteria isolated from companion animals in the Republic of North Macedonia", approved by the Faculty of Veterinary Medicine in Skopje (Decision No. 0202-359/11 from 31.3.2023).
The study had two phases. In the first, prospective phase, 48 isolates from clinical samples were analyzed for routine culture and bacteriology testing from April 2023 to May 2024. To ensure comprehensive data, in the second, retrospective phase, an additional 122 isolates from dogs were retrieved from the microbial strain collection of the Laboratory of Microbiology at the Faculty of Veterinary Medicine in Skopje (FVMS). These additional isolates were from clinical samples submitted between October 2019 and March 2023 for routine culture and bacteriology testing. All historical isolates were stored at -80 °C in 20% glycerol and tryptic soy broth (TSB; Oxoid, UK) and recultured before analysis. Importantly, no animals were specifically selected for participation in this study.
Samples included swabs from skin, nose, ears, eyes, vagina, and wounds/abscess swabs, milk, and urine samples. When appropriate, sampling sites were grouped into broader categories, such as combining skin samples with those from wounds and abscesses into a single category labeled "skin/soft tissue samples". All samples were cultured on 5% sheep blood agar (C-pharm, Croatia) and incubated at 37 °C for 24 h under aerobic conditions.Bacterial identification
Bacterial species were identified by culture morphology and MALDI-TOF MS (Bruker Daltonics, Bremen, Germany). For identification, a Direct Transfer Procedure (DT) was used. The addition of formic acid in the DT was used when necessary to ensure a reliable log (score). Measurements were performed using Flex Control 3.4 software, and results with a log (score) ≥2.0 were considered reliable and verified for species-level identification. Quality control was conducted using the reference strain). Measurements were performed using Flex Control 3.4 software, and results with a log (score) ≥2.0 were considered reliable and verified for species-level identification. Quality control was conducted using the reference strainStaphylococcus aureus ATCC 29213 to ensure accurate identification.
Antimicrobial susceptibility testing
Antimicrobial susceptibility was tested using the disc-diffusion method (Kirby-Bauer) with a panel of 12 antibiotics representing eight classes (Table 1). Oxacillin was used to screen for methicillin resistance in S. seudintermedius, and cefoxitin was used as a surrogate test as an indicator of methicillin resistance in coagulase-negative staphylococci (CoNS) and S. aureus . Antibiotic susceptibility interpretations followed CLSI guidelines: VETO1S-Ed6 (12) for canine-specific breakpoints and M100 standards ( 13 ) where species-specific breakpoints were unavailable (Table 1). To analyze the phenotypic resistance profiles, intermediate susceptibility results were categorized as resistant to account for their potential clinical significance.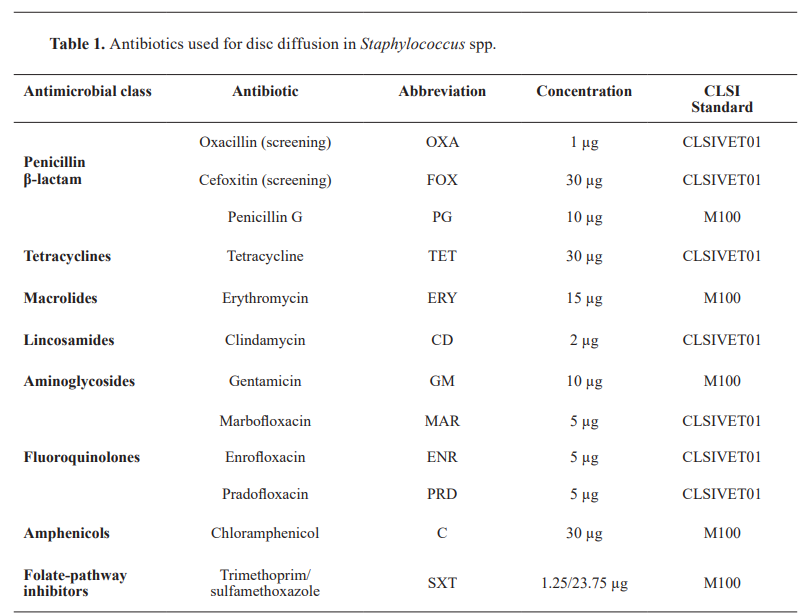
Molecular detection of resistance genes
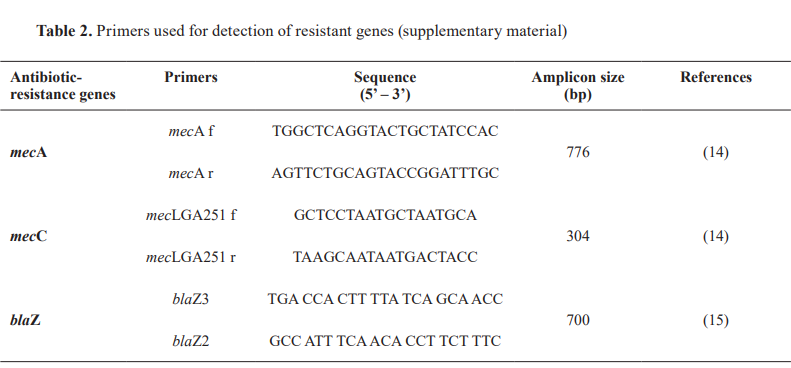
Bacterial DNA was extracted using the boiling method technique. One colony of a pure bacterial isolate was suspended with PBS solution (200 µl) and incubated in a thermoblock for 30 min at 95 °C. Conventional PCR was used to detect the mecA and mecC genes (14) in all isolates to identify methicillin resistance. Additionally, the blaZ gene (15) was tested in a subset of S. pseudintermedius isolates showing discordance between mec A results and oxacillin resistance. Specifically, 49 mecA- negative but oxacillin-resistant S. pseudintermedius isolates were analyzed for the presence of blaZ, as a determinant of beta-lactamase-mediated resistance. The primers and reaction conditions were the same as previously described (Table 2).
Statistical analysis
Associations between resistance patterns, bacterial species ( S. pseudintermedius , S. aureus, CoNS), and the presence of methicillin resistance genes (mecA, mecC, and blaZ) were analyzed using Fisher's Exact Test for contingency tables with expected cell frequencies below 5 and Pearson's Chi-square test for larger tables. Resistance and susceptibility rates were calculated for all isolates and specific subgroups (eg, methicillin-resistant strains), with 95% confidence intervals determined using the Wilson Score Interval method. The data was organized and analyzed in Microsoft Excel, and the statistical significance was defined as p<0.05.
RESULTS
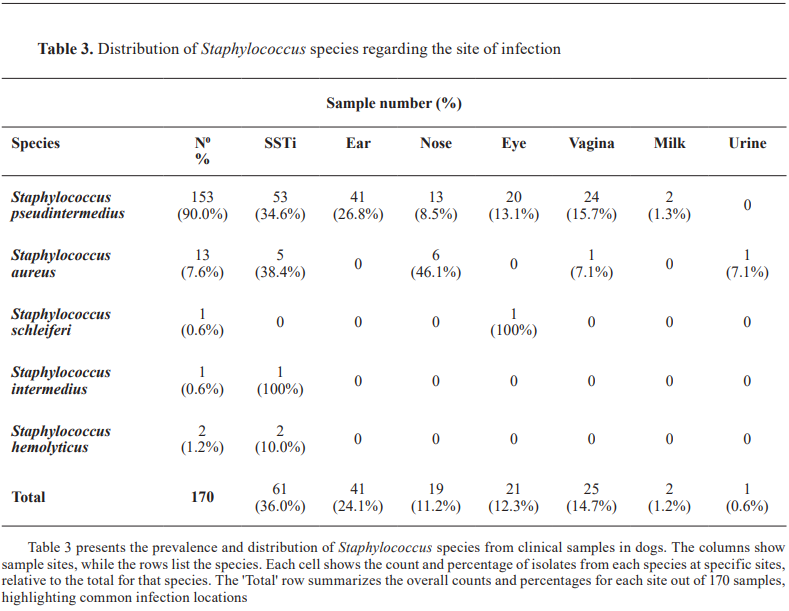
The study identified a total of 170 Staphylococcus isolates. The majority (153/170, 90%, 95% CI: 84.7–93.6%) were identified as Staphylococcus pseudintermedius, followed by Staphylococcus aureus (13/170, 7.6%, 95% CI: 4.2–12.7%), Staphylococcus hemolyticus (2/170, 1.2%, 95% CI: 0.2–4.3%), Staphylococcus aureus schleiferi (1/170, 0.6%, 95% CI: 0.03–3.50%), and Staphylococcus intermedius (1/170, 0.6%, 95% CI: 0.03–3.50%). The distribution of isolates across sample sites showed skin and soft tissue infections (SSTIs) as the most common source (61/170, 34.6%), followed by ear infections (41/170, 24.1%) and vaginal samples (25/170, 14.7%). Less frequent sources included ocular (21/170, 12.3%), nasal (19/170, 11.2%), milk (2/170, 1.2%), and urine samples (1/170, 0.6%) (Table 3).
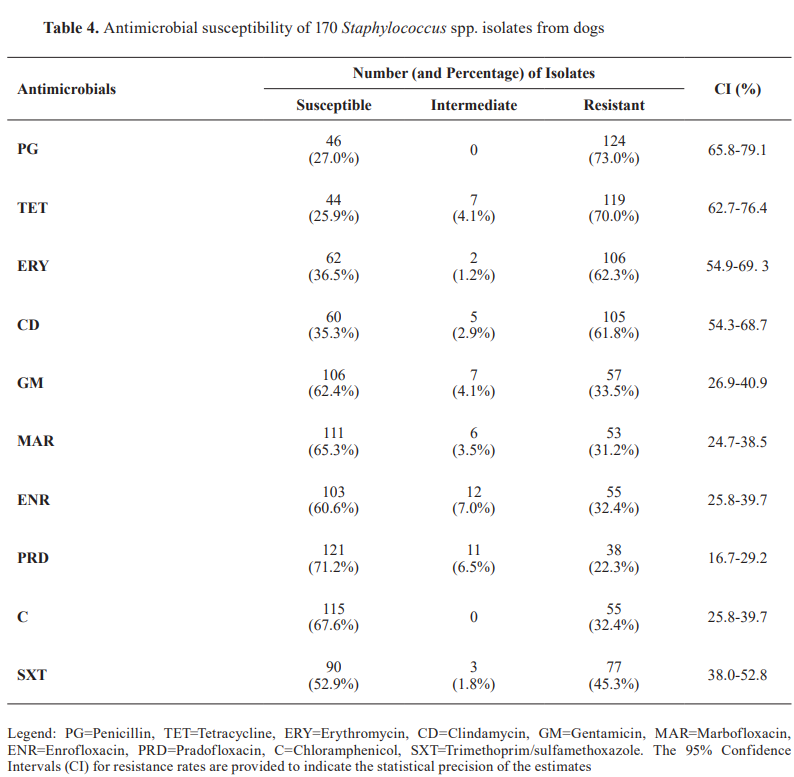
Antimicrobial resistance analysis revealed a significant variation across antibiotic classes (χ²=237.24, p<0.0001). Beta-lactams demonstrated the highest resistance rates, with 73% (124/170, 95% CI: 66.3–79.6%) of isolates resistant to penicillin G. Oxacillin/cefoxitin resistance was observed in 56.5% (96/170, 95% CI: 49.4–63.4%) of isolates. Tetracyclines showed 70% (119/170, 95% CI: 63.1–76.9%) resistance to tetracycline. Among macrolides and lincosamides, clindamycin resistance was 61.8% (105/170, 95% CI: 54.5–69.1%), while erythromycin resistance was 62.3% (106/170, 95% CI: 55.1–69.6%). Fluoroquinolones showed varied resistance: enrofloxacin resistance was 32.3% (55/170, 95% CI: 25.2–39.4%), marbofloxacin resistance was 31.2% (53/170, 95% CI: 24.2–38.2%), and the pradofloxacin resistance was the lowest at 22.3% (38/170, 95% CI: 16.0–28.6%). Among folate-pathway inhibitors, resistance to trimethoprim/sulfamethoxazole was 45.3% (77/170, 95% CI: 38.1–52.5%), while chloramphenicol resistance was 32.3% (55/170, 95% CI: 25.2–39.4%). ( Table 4 ).
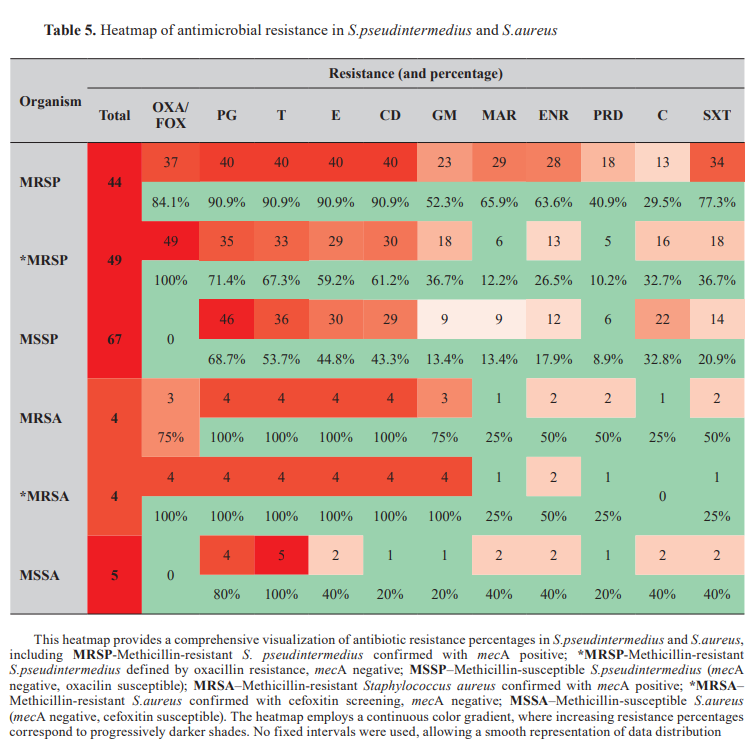
The antibiotic resistance patterns of S. pseudintermedius and S. aureus were analyzed and are summarized in Table 5. Multidrug resistance (MDR) prevalence across Staphylococcus species Multidrug resistance was observed in 70.5% of the isolates. Multidrug Resistance was observed in 69.2% of S. pseudintermedius isolates (106/153, 95% CI: 61.97%–76.59%) and 85% of S. aureus isolates (11/13, 95% CI: 54.55%–98.08%). Fisher's Exact Test revealed no statistically significant difference in MDR prevalence between species (p=0.134), likely due to the limited sample size of S. aureus isolates. Odds ratio analysis suggests that S. aureus isolates may be approximately 2.44 times more likely to exhibit MDR than S. pseudintermedius isolates (OR: 2.44). However, further validation with larger sample sizes is required.
The antibiotic resistance patterns of S.pseudintermedius and S.aureus were analyzed and are summarized in Table 5 . Multidrug resistance (MDR) prevalence across Staphylococcus species
Multidrug resistance was observed in 70.5% of the isolates (Table 6). Multidrug resistance was observed in 69.2% of S. pseudintermedius isolates (106/153, 95% CI: 61.97%–76.59%) and 85% of S. aureus isolates (11/13, 95% CI: 54.55%–98.08%). Fisher's Exact Test revealed no statistically significant difference in MDR prevalence between species (p=0.134), likely due to the limited sample size of S. aureus isolates. Odds ratio analysis suggests that S. aureus isolates may be approximately 2.44 times more likely to exhibit MDR than S. pseudintermedius isolates (OR: 2.44). However, further validation with larger sample sizes is required.
Oxacillin/Cefoxitin resistance
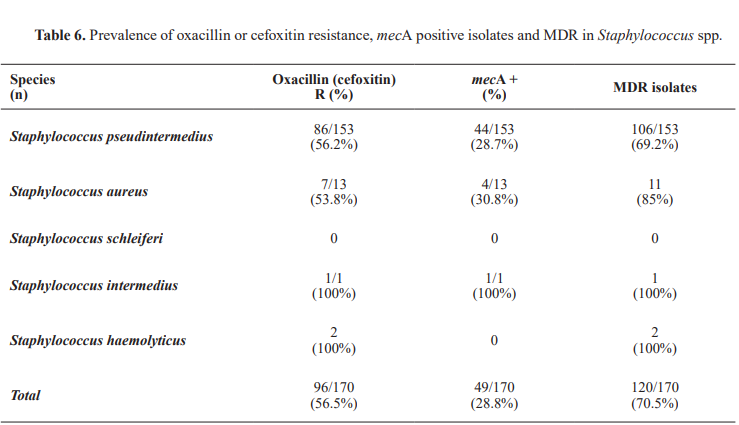
The mecA gene was detected in 28.8% of all Staphylococcus isolates (49/170,95%CI:23.0%–35.2%). Among the species, was identified in 28.7% of Staphylococcus pseudintermedius isolates (44/153, 95%CI: 22.4%–35.9%) and 30.8% of S.aureus isolates (4/13, 95%CI: 12.8%–58.6%). The mecC gene was not detected in any of the 170 isolates analyzed in this study.
Among the 153 Staphylococcus pseudintermedius isolates, phenotypic oxacillin resistance was observed in 86 isolates (56.2%; 95% CI: 48.1%–63.1%), which was nearly double from the prevalence of mecA.Fisher's Exact Test revealed a statistically significant difference between the prevalence of mecA and oxacillin resistance (p<0.001).
In Staphylococcus aureus (n=13), the discrepancy between prevalence (30.8%) and phenotypic cefoxitin resistance (53.8%) was not statistically significant (p=0.108), probably due to the small sample size.
Among the less commonly isolated species, the single S. intermedius isolate was oxacillin-resistant and mecA-positive. Both Staphylococcus haemolyticus isolates (2/2; 100%, 95% CI: 34.2%–100%) exhibited phenotypic cefoxitin resistance but lacked the mecA and mecC genes. The S. schleiferi isolates showed no resistance to the tested antimicrobials and did not carry the mecA or mecC genes.
Resistant profiles in Staphylococcus pseudintermedius Among the 153 S. pseudintermediusisolates, 145 (94.8%; 95% CI: 89.9%–97.8%) exhibited resistance to at least one antibiotic, while 8 isolates (5.2%; 95% CI: 2.2%–10.1%) showed no resistance to the antibiotics tested. Phenotypic oxacillin resistance was observed in 86 isolates (56.2%; 95% CI: 48.2%–63.8%), whereas it was detected in 44 isolates (28.7%; 95% CI: 22.4%–35.9%). Multidrug resistance was identified in 106 isolates (69.3%; 95% CI: 61.7%–76.1%). Among the 49 S. pseudintermedius isolates that were-negative but oxacillin-resistant, 32 isolates (65.3%; 95% CI: 51.0%–77.4%) were blaZ-positive, while 17 isolates (34.7%; 95% CI: 22.6%–49.0%) were blaZ negative. All blaZ-positive isolates were resistant to oxacillin, and 27 were resistant to penicillin G (84.3%; 95% CI: 61.4%–89.6%).
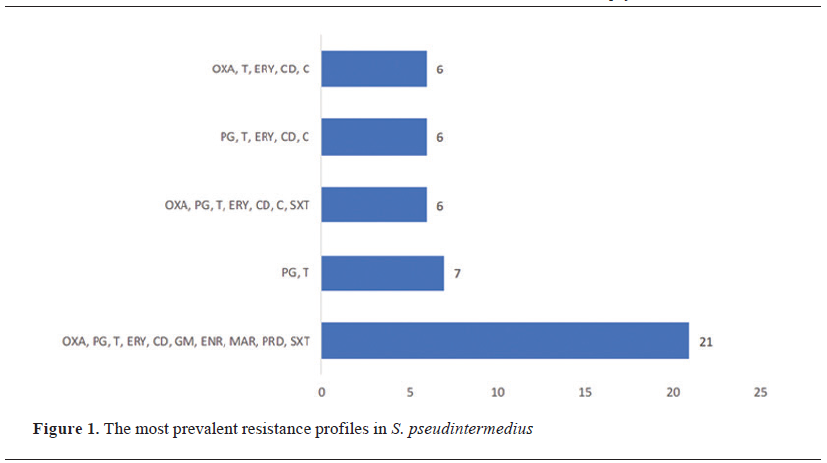
The analysis revealed 68 unique resistance profiles. Overall, the most frequent resistance profile (n=21) included OXA, PG, T, ERY, CD, GM, ENR, MAR, PRD, and SXT (Fig. 1). This profile was observed in 13 mecA-positive isolates (59.1%; 95% CI: 38.8%–76.7%) and 8 mecA-negative isolates (40.9%; 95% CI: 23.3%–61.2%) (Fig. 2). Further analysis revealed that all 8 mecA-negative isolates were blaZ-positive. One of the most frequent resistance profiles, observed in 6 isolates (12.2%; 95% CI: 5.7%–23.5%), included OXA, PG, T, ERY, CD, C, and SXT. Among these, 5 isolates were blaZ-positive (83.3%; 95% CI: 43.6%–97.0%), and 1 isolate was mecA-positive (16.7%; 95% CI: 3.0%–56.4%). A total resistance profile covering OXA, PG, T, ERY, CD, GM, ENR, MAR, PRD, C, and SXT was identified in 3 isolates, with 2 being mecA-positive (66.7%; 95% CI: 20.8%–93.9%) and 1 being blaZ-positive (33.3%; 95% CI: 6.1%–79.2%). (Fig. 2). 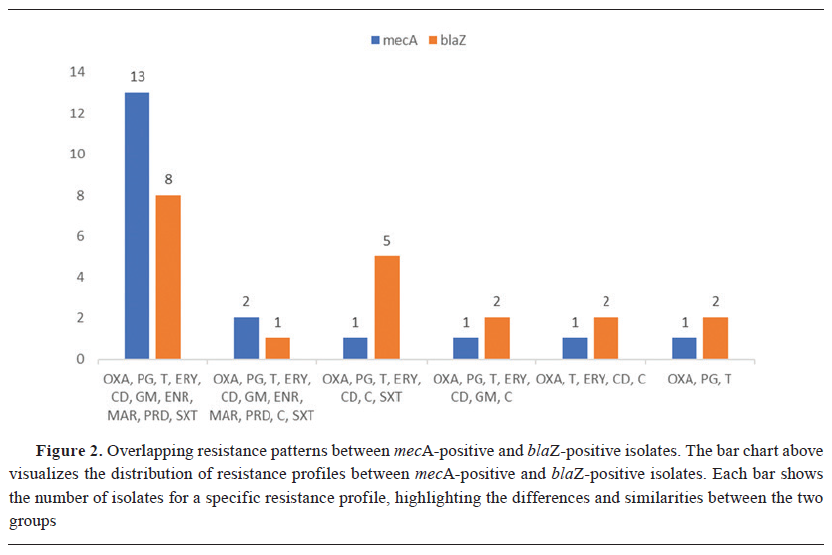
Resistant profiles in Staphylococcus aureus
All Staphylococcus aureus isolates exhibited resistance to at least one antibiotic. Of these, 4 isolates were mecA-positive (4/13, 36.4%, 95% CI: 14.9%–64.8%), and 7 were mecA-negative (7/13, 63.6%, 95% CI: 35.2%–85.1%).
The most common resistance profiles identified were FOX, PG, T, ERY, CD, GM (2 isolates: 1 mecA-positive, 50.0%, 95% CI: 9.5%–90.5%; 1 mecA-negative, 50.0%, 95% CI: 9.5%–90.5%) and FOX, PG, T, ERY, CD, GM, ENR, MAR, PRD, C, SXT (2 isolates: 1 mecA-positive, 50.0%, 95% CI: 9.5%–90.5%; 1 mecA-negative, 50.0%, 95% CI: 9.5%–90.5%). Unique resistance profiles were observed, including PG, T, ERY, ENR (1 isolate, 100%, 95%CI:21.7%–100%) and FOX, PG, T, ERY, CD, ENR, PRD (1 isolate, 100%, 95% CI: 21.7%–100%), mecA-negative isolates. A Chi-square test comparing resistance profiles between mecA-positive and mecA-negative isolates indicated a statistically significant difference (χ²=10.54, p=0.034). Both Staphylococcus hemolyticus isolates exhibited an identical resistance profile: FOX, PG, T, ERY, CD, GM, and S. Staphylococcus intermedius displayed the resistance profile: OXA, PG, T, ERY, CD, and SXT.
DISCUSSION
This study provides the first detailed analyzes of AMR profiles and genetic determinants in Staphylococcus spp. isolates from canine clinical samples in North Macedonia. These findings address a critical gap in AMR surveillance within the Balkan region, offering valuable insights into local resistance trends and their alignment with broader European patterns. The results demonstrate a high prevalence of MDR and MR among the isolates, underscoring the significant challenges posed by resistant strains. Specifically, 70.5% of isolates exhibited MDR, reflecting resistance to multiple antibiotic classes, including critically important antimicrobials (CIAs) for human medicine. The presence of mecA was confirmed in 28.8% of isolates, highlighting the growing threat of methicillin-resistant strains in companion animals.
Our study revealed a high prevalence of MRSP with 28.7% of isolates mecA-positive. These rates are significantly higher than those reported in Denmark, where oxacillin resistance ranges between 6-8%, and in Norway, which reports a 4.4% prevalence based on mecA detection (5). In the Balkan region, data on AMR is limited, with the prevalence of the mecA gene reported as 26.3% in Serbia (16) and 24.4% in Bosnia and Herzegovina (17). In contrast, Croatia reported a much lower prevalence, with only 7.5% of S. pseudintermedius isolates resistant to oxacillin (18). Additionally, Bulgaria reported a high prevalence of MDR in Staphylococcus spp. at 59.3% (19). Similarly, a study in Romania (20) reported that 82.8% of isolates exhibited MDR phenotypes, reflecting the widespread resistance challenges in the region. Our findings further emphasize the significant concern of MDR strains, with 69.3% of S. pseudintermedius and 85% of S. aureus isolates exhibiting resistance to multiple antimicrobial classes. The observed diversity in resistance rates across Europe highlights the multifaceted nature of AMR in Staphylococci isolated from dogs. This heterogenicity results from a combination of factors, including variability in antimicrobial prescribing practices and stewardship initiatives (10, 21). This evidence substantiates the necessity for cooperation in AMR surveillance, standardization of microbiological diagnostic methodologies, and implementation of antimicrobial stewardship initiatives to combat AMR effectively.
Methicillin resistance in S. pseudintermedius mecA and mecC genes. Interestingly, 27.6% of S. pseudintermedius (*MRSP) isolates showed phenotypic oxacillin resistance despite testing negative for the mecA and mecC genes. These findings are consistent with the study of Bertelloni et al. (6), which reported a similar discrepancy, with 44% of isolates phenotypically resistant to oxacillin and only 24% positive for mecA. Such discrepancies highlight the complexity of methicillin resistance mechanisms in Staphylococcus spp., including the potential involvement of alternative genetic determinants. Furthermore, our study highlights the critical role of beta-lactamase-mediated resistance, with blaZ detected in 65.3% of *MRSP isolates. This suggests that hyperproduction of beta-lactamase may contribute to oxacillin resistance in mecA-negative isolates (22, 23, 24). This finding aligns with the study of Arede et al. (25) who demonstrated the significant role of blaZ in resistance expression in MRSA. This finding emphasizes the importance of extending diagnostic testing to include blaZ detection, especially in regions where mecA-negative oxacillin resistance is prevalent.
Beyond resistance to β-lactams, MR isolates show co-resistance to multiple antibiotic classes, including tetracyclines, macrolides, lincosamides, and fluoroquinolones (6, 26). This study identified a significant correlation between MR resistance to aminoglycosides, fluoroquinolones, lincosamides, and macrolides, emphasizing the frequent occurrence of co-resistance in MR strains (27, 28). A concerning finding in this study is the widespread multidrug resistance exhibited by isolates. In particular, S. pseudintermedius showed remarkable resistance patterns: three isolates (3.2%) were resistant to all eight antibiotic classes tested, while twenty-one isolates (13.7%) were resistant to seven: oxacillin, penicillin G, tetracycline, erythromycin, clindamycin, gentamicin, enrofloxacin, marbofloxacin, pradofloxacin, and trimethoprim-sulfamethoxazole. This is consistent with the pattern reported by Morais et al. (27), where 30.4% of isolates showed resistance to beta-lactams, erythromycin, clindamycin, tetracyclines, gentamicin, fluoroquinolones and trimethoprim-sulfamethoxazole.
Additionally, two isolates of S. aureus (18.2%) were resistant to all antimicrobials tested, highlighting therobust adaptability of S. aureus as a species. Furthermore, the high prevalence of multidrug resistance (MDR) in S. aureus, with 84.6% exhibiting resistance to multiple drug classes, raises serious concerns regarding treatment options and highlights the critical need for effective antimicrobial stewardship.
The high rate of resistance to clindamycin, alincosamide classified as category C (29), is a significant concern in veterinary medicine. Clindamycin is widely recommended as one of the first-line antibiotics for treating skin infections in dogs (4, 30). However, this study found a resistance rate of 61.8% in Staphylococcus isolates, which significantly compromises its efficacy as a primary therapeutic option. The increasing resistance to clindamycin not only reduces its clinical use, but also limits available treatment options for common infections, potentially leading to the overuse of broader-spectrum or critically important antibiotics, such as fluoroquinolones.
Equally concerning are the resistance rates to fluoroquinolones, with 31.2% for marbofloxacin and 32.3% for enrofloxacin. As CIAs, fluoroquinolones play a crucial role in the treatment of serious infections in veterinary and human medicine. Their widespread use in small animal practice in our country (31) likely contributes to the observed resistance and emphasizes the need to limit their use to cases where alternatives have failed and susceptibility data demonstrate their efficacy. In contrast, pradofloxacin had the lowest resistance rate among the antibiotics tested, with only 22.3% of isolates showing resistance. This result is in line with our previous study (31), which highlighted the limited use of pradofloxacin in small animal practice. These lower resistance levels are likely due to the fact that pradofloxacin has only recently been introduced in the country and was approved in 2021 (32).
These findings emphasize the need to revise empirical treatment protocols and prioritize antimicrobial susceptibility testing (AST) to guide therapy. Our prior study (31) revealed that veterinarians primarily rely on "scientific literature" (45.61%) and "personal experience" (43.86%) when selecting antimicrobials for treatment. While these factors contribute to informed decision-making, they may also lead to variability in prescribing practices, particularly in regions lacking robust local resistance data (10). To address these challenges, promoting AST and providing veterinarians with region-specific resistance data are critical steps towards optimizing antimicrobial use and reducing resistance (28).
While this study provides valuable insights into MR and MDR S. pseudintermedius and S. aureus in North Macedonia , several limitations must be acknowledged. First, the study population was derived from diagnostic samples, which may overrepresent resistant strains, as veterinarians tend to submit samples primarily from treatment failures or recurrent infections (31). Furthermore, the lack of differentiation between first infections and those previously treated with antibiotics introduces variability in the dataset, potentially biasing the results (33). Despite these limitations, the findings highlight the urgent need for antimicrobial stewardship. Future research should include molecular tools like MLST to better understand resistance mechanisms and transmission dynamics in North Macedonia.
CONCLUSION
In conclusion, this study reveals a significant prevalence of methicillin resistance and multidrug resistance in Staphylococcus spp. from canine clinical samples in North Macedonia. The notable resistance of Staphylococcus pseudintermedius and Staphylococcus aureus to critical antimicrobials raises concerns about treatment efficacy. The detection of blaZ in mecA-negative isolates underscores the complexity of resistance mechanisms and highlights the need for molecular diagnostics in routine antimicrobial resistance surveillance. These findings emphasize the urgent need for antimicrobial stewardship and targeted AMR strategies to address the spread of resistant strains while understanding their clonal distribution to inform effective control measures.
CONFLICT OF INTEREST
The authors declare that they have no financial or non-financial conflict of interest regarding authorship and publication of this article.
ACKNOWLEDGMENTS
The authors acknowledge that the research was made through financial support from the Faculty of Veterinary Medicine in Skopje, project "Antimicrobial resistance in bacteria isolated from companion animals in the Republic of North Macedonia" (FVMS-IPR-4), for which they express their deepest gratitude.
AUTHORS' CONTRIBUTION
IC conceived the study and supervised the manuscript's writing. IS drafted the original manuscript, analyzed the data, and interpreted the results. ZPH performed the PCR analysis. IM, MJP, and MRM participated in reviewing and editing the manuscript. AC was involved in manuscript writing and reviewing. All authors have reviewed and approved the final version of the manuscript.

 10.2478/macvetrev-2025-0022
10.2478/macvetrev-2025-0022